Figures & data
Figure 1. Offspring exposed to maternal obesity and diabetes have a latent predisposition to metabolic disease, which is revealed by a high-fat Western-style diet. (A) Experimental strategy. (B) Body weights of male and female offspring in each of the four groups in (A) (male: LnC n = 14, ObC n = 13, LnHF n = 15, ObHF n = 9; female: LnC n = 19, ObC n = 13, LnHF n = 13, ObHF n = 18); inset, pre-weaning weights for Lean-born and Obese-born animals. (C) Blood glucose levels in male and female animals during a glucose tolerance test at 6 weeks of age (male: LnC n = 14, ObC n = 13, LnHF n = 15, ObHF n = 9; female: LnC n = 20, ObC n = 6, LnHF n = 13, ObHF n = 11). (D) Blood glucose levels in animals during an insulin tolerance test at 7 weeks of age (male: LnC n = 14, ObC n = 17, LnHF n = 19, ObHF n = 19; female: LnC n = 20, ObC n = 13, LnHF n = 13, ObHF n = 18). Data are represented as mean ± SEM. Statistically significant differences are as indicated: *p < 0.05, **p < 0.01, ***p < 0.001 (LnC vs ObC); †p < 0.05, ††p < 0.01, †††p < 0.001 (LnHF vs ObHF).
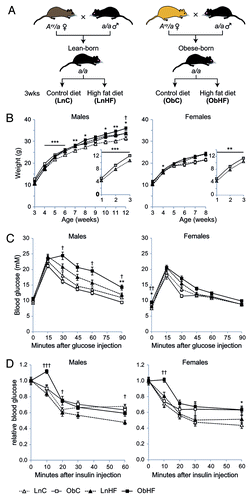
Figure 2. Offspring exposed to maternal obesity and diabetes have defects in lipid metabolism which are exacerbated by a Western diet. Total liver (A) triacylglycerol and (B) diacylglycerol levels (LnC n = 4, ObC n = 6, LnHF n = 12, ObHF n = 8), and (C) steatosis scores (LnC n = 4, ObC n = 9, LnHF n = 10, ObHF n = 10) in livers of 12 week old male animals. Representative liver histology sections are shown in (D) (scale bar, 100 μm). (E) Serum leptin levels and (F) relationship between serum leptin levels and body weight at 12 weeks (LnC n = 6, ObC n = 7, LnHF n = 12, ObHF n = 12). Box and whisker plots show the median, 25th and 75th percentile values with whiskers indicating the maximum and minimum; means are indicated by an X. Statistically significant differences are indicated by asterisks: *p < 0.05, **p < 0.01, ***p < 0.001. Right-tailed Fisher’s exact test, LnHF c.f. ObHF steatosis scores: p = 0.012.
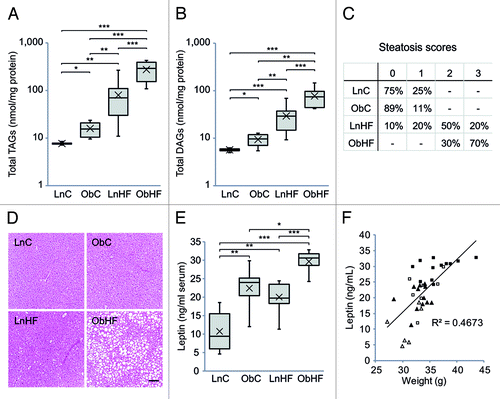
Figure 3. The latent metabolic phenotype is associated with changes in hepatic gene expression. (A) Heat map of gene transcripts with significant expression changes in ObC vs. LnC offspring (n = 4 per group); note the similar patterning across all animals exposed to overnutrition (prenatally, postnatally or both). The columns on the right indicate genes that have CpG island promoters (green) or do not (white), and whether the gene was identified by LimmaGP (white), Messina (black) or both (gray). Average log2-fold expression changes are shown at the far right. (B) Bar graph showing –log p-values (with Benjamini–Hochberg correction for multiple testing) for gene ontologies significantly overrepresented in genes that are differentially expressed in ObC animals relative to LnC. A p-value cutoff of 0.05 is indicated by the red line. (C) Bar graph showing relative copy number of mitochondrial genes Atp6 and Nd6 (relative to the nuclear gene Ndufv1) in livers of LnC, ObC, LnHF and ObHF animals. Data are represented as mean ± SEM. Statistically significant differences are indicated by asterisks: *p < 0.05, **p < 0.01, ***p < 0.001.
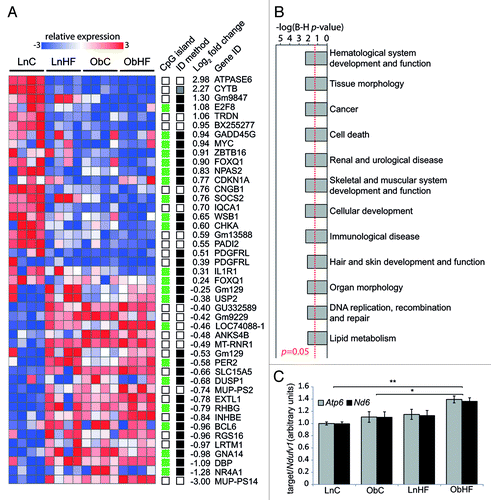
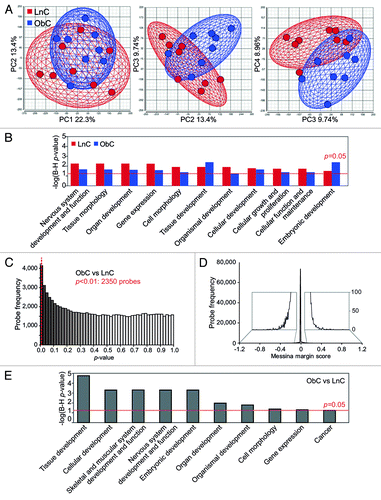
Figure 4. Livers of obese-born males exhibit widespread changes in cytosine methylation. (A) Two-dimensional plots showing principal component (PC) scores of LnC and ObC animals (n = 8 per group). Three plots are shown: scores for PC 1 vs. PC2 (left), PC2 vs. PC 3 (middle) and PC 3 vs. PC 4 (right). The amount of variability accounted for by each PC is indicated on the axes. The wireframe ellipses surrounding PC scores for each group are indicative of the overall standard deviation within each group. (B) Bar graph showing –log p-values (with Benjamini–Hochberg correction for multiple testing) for gene ontologies significantly overrepresented in methylation variable genes in LnC and ObC animals. A p-value cutoff of 0.05 is indicated by the red dashed line. (C) Histogram showing the distribution of LimmaGP p-values across all probes on the methylation array. A p-value cutoff of 0.01 is indicated by the red dashed line. (D) Histogram showing the distribution of Messina margin scores from the methylation array. Inset shows enlarged view of the histogram tails. (E) Bar graph showing –log p-values (with Benjamini-Hochberg correction for multiple testing) for gene ontologies identified as being significantly overrepresented in genes differentially methylated in ObC vs LnC (as detected by LimmaGP, p < 0.01). A p-value cutoff of 0.05 is indicated by the red dashed line.