Figures & data
Figure 1. Schematic physical maps of human (top) and mouse (bottom) orthologous imprinted GPR1-GPR1AS-ZDBF2/Gpr1-Zdbf2linc-Zdbf2 clusters. Imprinted genes are represented as follows: Open and filled boxes represent untranslated regions and ORFs, respectively; arrows indicate transcription orientation. Human GPR1AS does not contain any significant ORFs. ZDBF2_v2 codes for an ORF similar to the original ZDBF2 protein sequence encoded by ZDBF2_v1 (see Fig. S1). Magenta and cyan bars represent maternally or paternally methylated DMRs in mice. The extent of each DMR was determined in previous DNA methylome studies.Citation20-Citation22
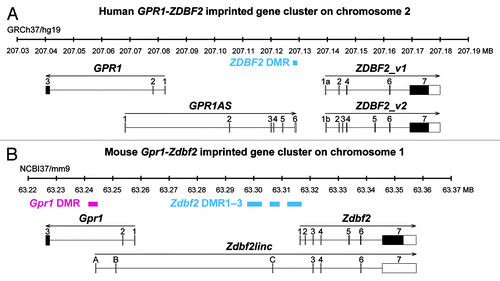
Figure 2. Relative expression of GPR1AS/Zdbf2linc in multiple human and mouse tissues (qRT-PCR). Expression in the human placenta (Family ID: 055) or mouse placenta at E15.5 is set as 1 for each gene. UD means undetectable after 40 cycles of amplification.
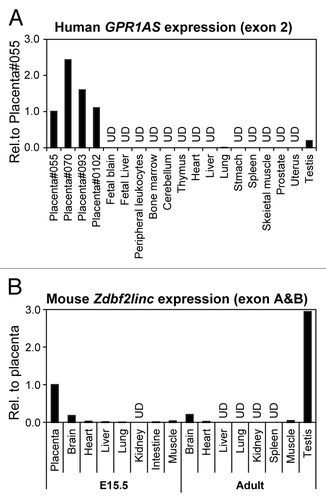
Figure 3. Allele-specific RT-PCR sequencing of GPR1AS and ZDBF2 in human placentas. Four heterozygous genotypes were used to distinguish between maternal and paternal alleles in human placental samples from four Japanese patients (National Center for Child Health and Development). SNP positions are highlighted in yellow.
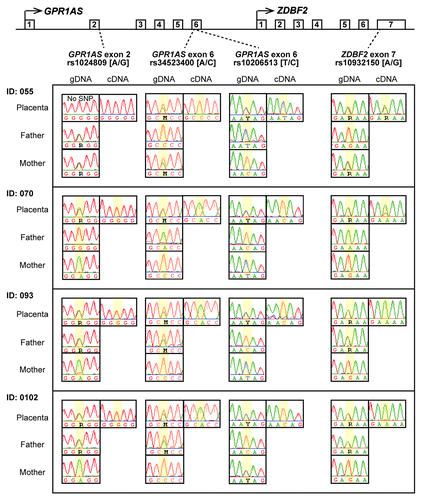
Figure 4. Bisulfite sequencing of the GPR1AS promoter region in human placenta and umbilical blood samples. Each row represents the results of an independent sequencing reaction. Open and closed circles denote unmethylated and methylated CpGs. Calculated methylation levels are shown below each graph. Heterozygous SNP rs1683074 was used to distinguish between maternal and paternal alleles in human samples. One of case (Family ID: 055) was also investigated by allelic expression analysis in .
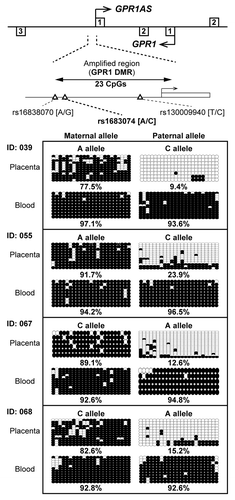
Figure 5. (A) Embryonic and extra-embryonic expression of Zdbf2linc in pre- and post-gastrulation mouse embryo (at E3.5 to E9.5). Abbreviations: E3.5 Blast, pools of 5 blastocysts (gray bars); E5.5 Emb, whole embryos (gray bars), E6.5-E9.5, dissected embryos free of extra-embryonic tissues (black bars); E6.5-E7.5 Exe, extra-embryonic tissues (white bars); E9.5 Epc, ectoplacental cones dissected from the remaining extra-embryonic tissues (white bars). After normalization against the housekeeping Actb gene, relative expression in mouse blast #1 is set as 1. UD indicates undetected mRNA expression. (B) Allele-specific RT-PCR sequencing of Zdbf2linc-specific exon (left) and common Zdbf2 exon (right) in mouse ectoplacental cones at E9.5 and placentas at E15.5. We prepared 2 parental mouse strains, C57BL/6N (B6) and JF1/Msf (JF), and reciprocal F1 hybrids (BJF1; F1 hybrid derived from mating between B6 female and JF male mice, JBF1; the reciprocal cross of JF female with B6 male mice)
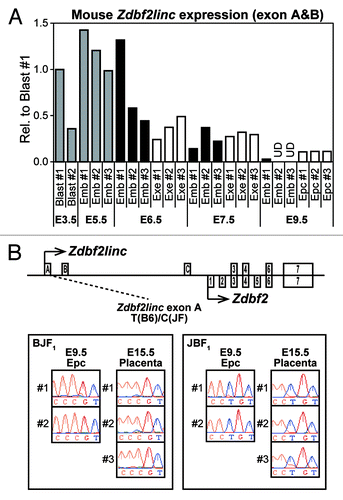
Figure 6. Bisulfite sequencing of Gpr1 DMR in mouse BJF1/JBF1 embryos and placental tissues at E9.5. M and P indicate maternally and paternally inherited alleles. Both alleles were discriminated by more than one polymorphism.
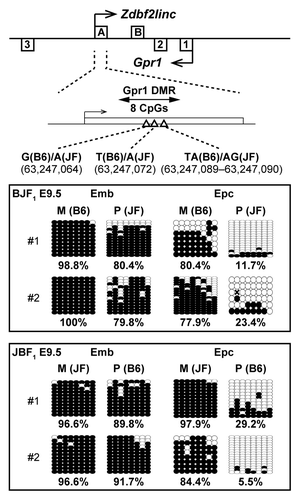
Figure 7. Bisulfite sequencing of ZDBF2 DMR and ZDBF2 promoter in human placenta and umbilical blood samples. Calculated methylation levels are shown below the graphs. Heterozygous SNP rs10206513 was used to distinguish between maternal and paternal alleles in human samples (all cases were tested in ).
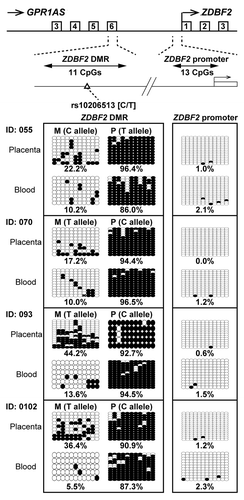
Figure 8. Human and mouse sequences of the imprinted GPR1-ZDBF2 domain. Percent Identity Plot (PIP, a scale of 50% to 100% conservation, on the y-axis) showing order and alignment of the entire imprinted domain on human chromosome 2q33.3 (chr2: 207,030,001–207,190,000, on the x-axis) and the orthologous region on mouse chromosome 1C2. Detected sequence homology (>50% identity) is plotted by position along the test sequence and level of sequence similarity. Structural features in the human ortholog, including exons, repeats and CpG-rich regions, are shown above the top line. The novel imprinted lncRNA gene, GPR1AS, is shown in blue, and known imprinted protein-coding genes, GPR1 and ZDBF2, are shown in green.
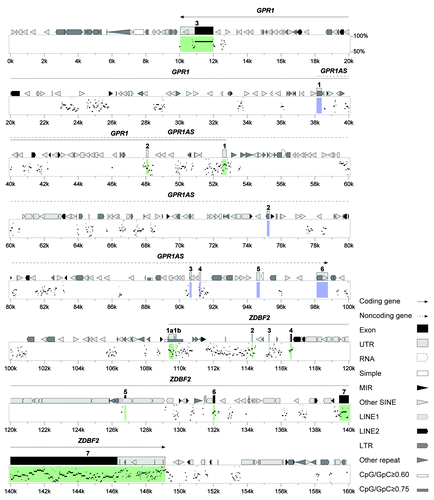
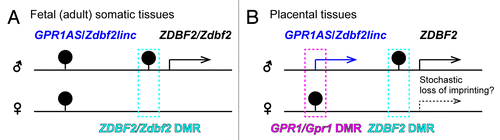
Figure 9. Summary of allele-specific epigenetic and gene expression differences within the GPR1AS-ZDBF2 (Zdbf2linc-Zdbf2) locus during fetal (A) and extra-embryonic (B) development. Arrows represent transcriptional orientation of individual imprinted genes. DMR positions are indicated by filled pins on the methylated alleles.