Figures & data
Figure 1. Breast cancer cell line radioresistance. Clonogenic survival of the MDA-MB-231 breast cancer cell line was performed to determine relative radioresistance.
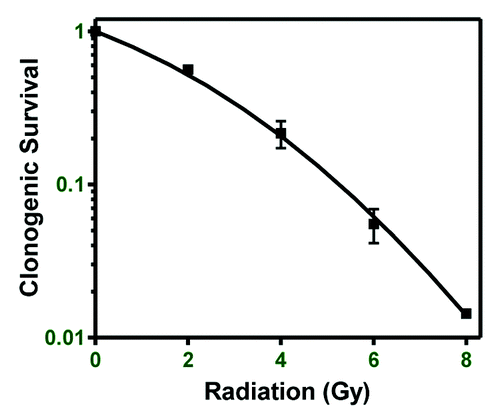
Figure 2. Global and specific changes in DNA methylation. Global DNA methylation changes were determined in MDA-MB-231 cells following 2 and 6 Gy. (A) The number of loci hypomethylated or hypermethylated 1–72 h post-irradiation with 2 (left) or 6 Gy (right). (B) The number of genes displaying differential DNA methylation 1–72 h post-irradiation and the number of common differentially methylated genes between the 2 and 6 Gy dose at each time point. (C) Differential DNA methylation and gene expression at 1 h post-2 Gy in 3 chosen genes. (D) Differential DNA methylation in the RB1 gene 1–72 h post-2 Gy correlated with gene expression. (E) Measurement of RB1 gene methylation by meDiP-qPCR 1–72 h post-2 Gy. Values are relative to matched 0 Gy controls.
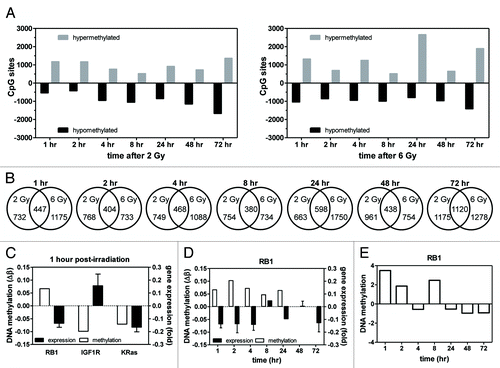
Table 1. Common differentially methylated genes
Figure 3. DNMT1 expression following irradiation. DNMT1 expression was determined in MDA-MB-231 cells 1–72 h after treatment with 0, 2, or 6 Gy. Decreased DNMT1 expression is observed following 6 Gy.
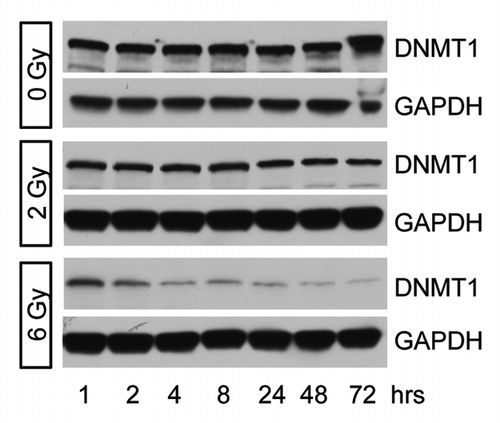
Figure 4. DNA methylation of cell cycle pathways. Gene ontology analysis was used to determine that cell cycle-related pathways are enriched for DNA methylated loci and this was compared with biological response. (A) The average DNA methylation state of genes in the indicated pathways at 1–72 h post-irradiation. Blue boxes indicate hypomethylation; red boxes indicate hypermethylation. The absence of a box indicates this pathway was not significantly enriched at that time/dose. (B) Cells were stained with propidium iodide to determine cell cycle. Representative histograms at 3 time points are shown. Numbers shown represent the percentage in G2/M. (C) Time course of the percentage of cells in G2/M following irradiation (n = 3). The percentage of cells in G2 phase approached control levels in 2 Gy-treated cells by 72 h post-irradiation, but a high percentage of G2 cells remained 72 h post-6 Gy.
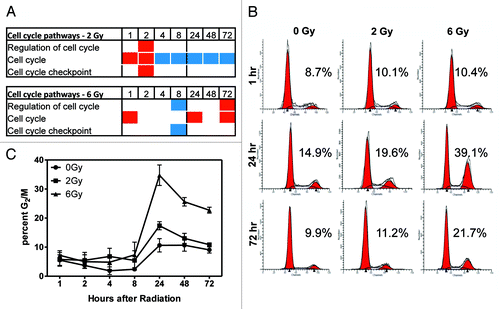
Table 2. DAVID Functional Annotation Chart results
Figure 5. Gene ontology analysis of DNA repair pathways and the time course of DNA repair. (A) Gene ontology analysis showed enrichment of differentially methylated genes in DNA repair pathways. Red boxes indicate the average delta-β of genes differentially methylated at that condition and represent hypermethylation. Blue boxes indicate hypomethylation. The absence of a box indicates that pathway was not enriched at that condition. (B) Analysis of DNA damage and repair by γH2AX immunofluorescence. γH2AX is in green and blue is Dapi staining of nuclei. Shown are three representative time points. (C) Quantitation of γH2AX foci across the full time range (n = 3). (D) western blot showing γH2AX staining following irradiation (representative of n = 3). DNA damage was greatest 1 h post-irradiation and decreased to basal levels in 2 Gy samples. Some staining remained >24 h post-6 Gy.
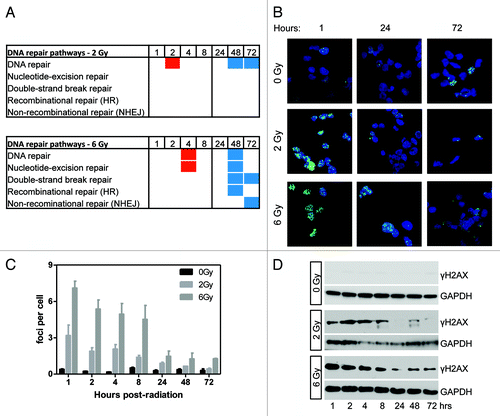
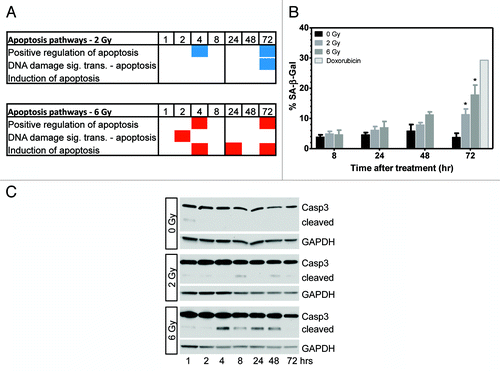
Figure 6. Gene ontology analysis shows enrichment of apoptosis pathways following irradiation. (A) Differentially methylated genes were enriched in apoptosis pathways following 2 and 6 Gy. Red and blue boxes indicate genes in the pathway were hypermethylated or hypomethylated, respectively. (B) Analysis of cellular senescence 8–72 h post-irradiation. Significant senescence was observed at 72 h post-irradiation (P < 0.05, n = 3). (C) Determination of apoptosis by western blotting for cleavage of caspase 3 (representative of n = 3). Low levels of cleaved caspase 3 were seen 4–48 h post-6 Gy.