Figures & data
Table 1. Data from microarray performed on E2f6 wild-type and E2f6 knockout MEFs
Table 2. CGI class definition of Stag3, Smc1β, Tuba3a, and Slc25a31 according to the more stringent criteria proposed by Takai and JonesCitation17
Figure 1. Deletion of E2f6 disrupts somatic cell CGI methylation at E2f6 dependent germline specific gene promoters. (A, C, E, and G) Bisulfite DNA sequencing analysis for CGI methylation of the Stag3, Smc1β, Tuba3a, and Slc25a31 promoters, respectively, in wild-type and E2f6−/− MEFs. Each gene representation includes the first exon in black and the position of the CGI in gray. Short lines with numbers depict the first nucleotide from the forward and reverse primers used for bisulfite sequencing. Percent methylation for the region amplified by the primers is given in parenthesis. (B, D, F, and H) Quantification of bisulfite sequencing data demonstrating the dynamics of methylation at each individual CpG dinucleotide in Stag3, Smc1β, Tuba3a, and Slc25a31 promoters, respectively. A value of one corresponds to 100% methylation. *Designates the position of E2f6 binding; +1 designates the TSS.
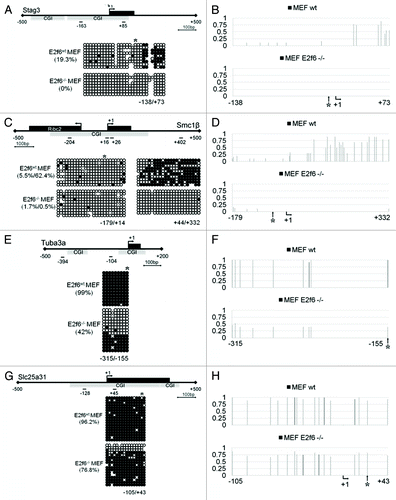
Table 3. Data from microarray performed on ESCs, EpiSCs, and MEFs
Figure 2. Germline-specific genes regulated by E2f6 are first silenced in primed pluripotent stem cells. (A) RNA expression levels of Stag3, Smc1β, Tuba3a, and Slc25a31 in ESCs, EpiSCs, and MEFs shown by qPCR. The expression of each gene in embryonic stem cells was set to a value of one. (B) ChIP-qPCR analysis for H3K4me3 enrichment spanning 2.5 kb downstream and 3 kb upstream (relative to the TSS) of the Stag3 promoter in ESCs, EpiSCs, and MEFs. Enrichment is shown normalized to the total H3 content at each specific primer pair position. (C) ChIP-qPCR analysis for enrichment of E2f6 at the Stag3 promoter in ESCs, EpiSCs, and MEFs. Enrichment is shown as the percent of Input DNA.
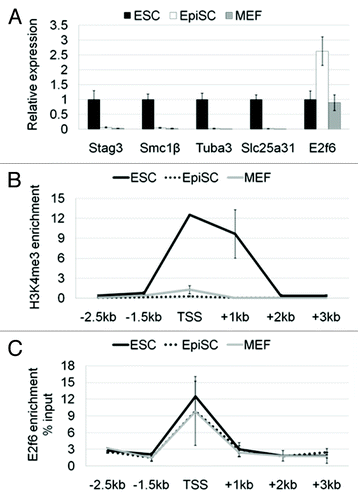
Figure 3. Differential DNA methylation of the Stag3 and Smc1β promoters in ESCs, EpiSCs, and MEFs. (A) CGI methylation of the Stag3 promoter in ESCs, EpiSCs, and MEFs. (B) CGI methylation of the Smc1β promoter in ESCs, EpiSCs, and MEFs. *Designates the position of E2f6 binding site; +1 designates the TSS.
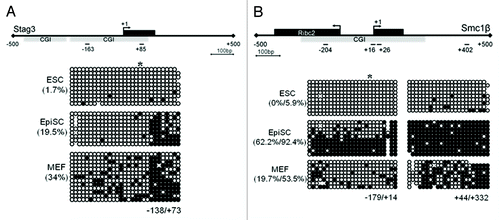
Figure 4. The effects of E2f6 and Dnmt3b overexpression on RNA expression and DNA methylation of meiotic genes in ESCs. (A) qPCR analysis of Stag3, Smc1β, and E2f6 RNA expression in ESCs with or without stable overexpression of E2f6. (B) Bisulfite DNA sequencing analysis of the Stag3 promoter in ESCs with or without stable overexpression of E2f6. (C) ChIP-qPCR analysis for E2f6 occupancy at the Stag3 promoter in ESCs ectopically overexpressing FLAG-tagged E2f6. (D) RNA expression levels of Stag3, Smc1β, and Dnmt3b in ESCs with or without stable overexpression of Dnmt3b. (E) Bisulfite DNA sequencing analysis for methylation of the Stag3 promoter in ESCs with or without stable overexpression of Dnmt3b.
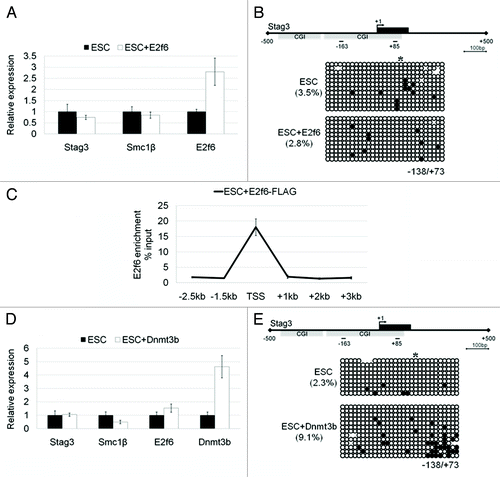
Figure 5. Downregulation of Stag3 and Smc1β gene expression during embryoid body (EB) differentiation of wild-type, Dnmt3b knockout (KO), and Dnmt3a/3b double knockout (DKO) J1 ESCs. (A) qPCR analysis of Stag3 and Smc1β RNA expression during EB differentiation of wild type, 3b KO, and 3a/3b DKO J1 ESCs at Day0 and Day 10. (B) CGI methylation of the downstream region of the Smc1β promoter measured using bisulfite sequencing from the sample of Day 0 and Day 10 following EB differentiation of wild-type J1 ESCs, Dnmt3b KO (C), and Dnmt3a/3b DKO (D).
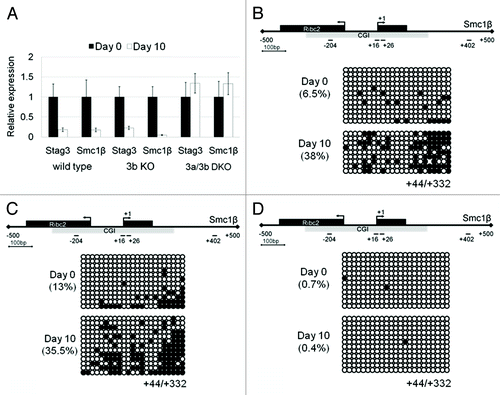
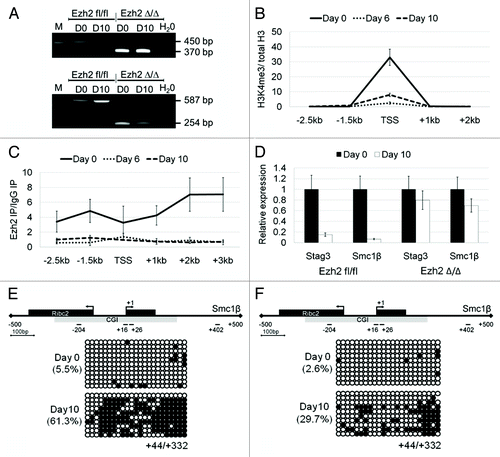
Figure 6. Downregulation of Stag3 and Smc1β gene expression during embryoid body (EB) differentiation of wild-type and Ezh2-deficient pluripotent stem cells. (A) Confirmation of Ezh2fl/fl and Ezh2∆/∆ genotype using PCR and RT-PCR, upper and lower panel, respectively. The amplicons for the flox and the Δ allele are 450 bp and 370 bp (for genomic PCR) and 587 bp and 254 bp (for cDNA PCR), respectively. (B) ChIP-qPCR analysis for H3K4me3 enrichment of the Stag3 promoter in Ezh2 fl/fl iPSCs at 3 differentiation time points (Day 0, Day 6, and Day 10). Enrichment is shown normalized to the total H3 content at each specific primer pair position. (C) ChIP-qPCR analysis for Ezh2 occupancy at the Stag3 promoter in Ezh2 fl/fl iPSCs at 3 differentiation time points (Day 0, Day 6, and Day 10). Enrichment is shown normalized to the background from a ChIP with mouse IgG. (D) qPCR analysis of Stag3 and Smc1β RNA expression during EB differentiation of Ezh2fl/fl iPSCs and Ezh2∆/∆ iPSCs at Day 0 and Day 10. The expression of each gene at Day 0 was set to a value of one. (E and F) CGI methylation of the downstream region of the Smc1β promoter measured using bisulfite sequencing from the sample of Day 0 and Day 10 following EB differentiation of Ezh2fl/fl iPSCs (E) and Ezh2∆/∆ iPSCs (F).