Figures & data
Figure 1. Expression of eNOS and arginase-2 in normal and IUGR cells relative to normal HUAEC. Quantification of eNOS (A) and arginase-2 (B) mRNA levels in control (open bars, n = 5) and IUGR (solid bars, n = 5) cultured HUAEC and HUVEC. Values are mean ± SEM *p < 0.05, vs. corresponding normal cells, #p < 0.05 vs. control HUAEC, one-way ANOVA.
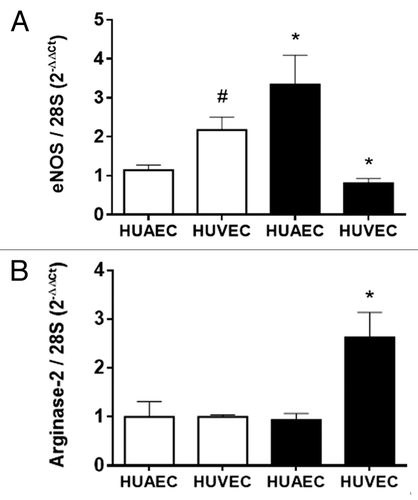
Figure 2. DNA methylation pattern in the NOS3 promoter of placental and umbilical endothelial cells. (A) Regions analyzed included the core promoter (-100 to -51), proximal promoter (-361 and -352), the shear stress response element (-992 to -976) and the hypoxia response element (-5375 and -5369). Specific CpG methylation (%) determined by pyrosequencing of bisulfite-treated DNA from HUAEC (white bars, n = 9), PLAEC (light gray bars, n = 4) and HUVEC (dark gray bars, n = 9) isolated from control (B) and IUGR (C) placentae. Values are mean ± SEM *p < 0.05, vs. HUAEC, two-way ANOVA.
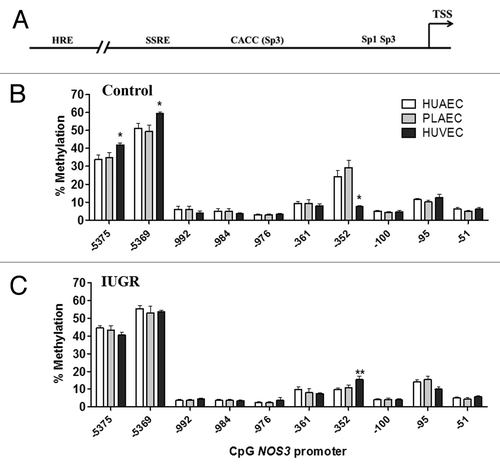
Figure 3. Comparison of DNA methylation status in the NOS3 promoter of normal and IUGR cells. Specific CpG methylation (%) in normal and IUGR-HUAEC (A), PLAEC (B) and HUVEC (C) determined by pyrosequencing of bisulfite-treated DNA samples. Values are mean ± SEM ***p < 0.001, *p < 0.05, vs. corresponding control EC, two-way ANOVA.
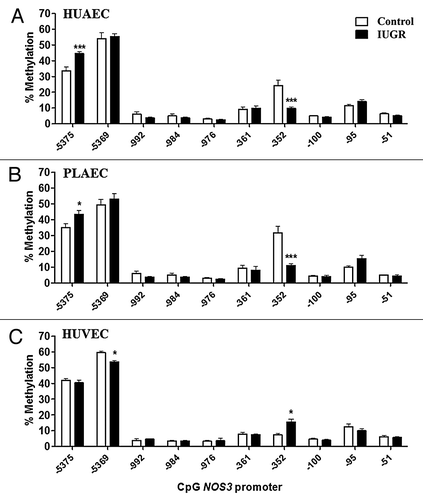
Figure 4. Prediction of transcription factors binding sites in ARG2 promoter. Cognate sequences for human transcription factors (underlined) and CpGs (highlighted in yellow) in the proximal and core promoter of ARG2 gene. Proposed transcription factor were obtained by in silico analysis of ARG2 promoter sequence with MatInspector software, and CpGs were numbered according to the position regarding the reported ATG.
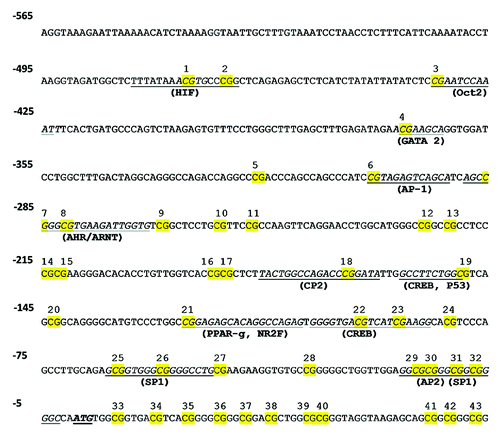
Figure 5. DNA methylation status in the ARG2 promoter of placental and umbilical endothelial cells. (A) Regions analyzed included the core (-33 to -7) and proximal (-471 and -281) promoter, where putative transcription factors or consensus binding sites are indicated. Specific CpG methylation (%) was determined by pyrosequencing of bisulfite-treated DNA from HUAEC (white bars, n = 9), PLAEC (light gray bars, n = 4) and HUVEC (dark gray bars, n = 9) isolated from normal (B) and IUGR (C) placentae. Values are mean ± SEM *p < 0.05 vs. HUAEC, #p < 0.05 vs. PLAEC, two-way ANOVA.
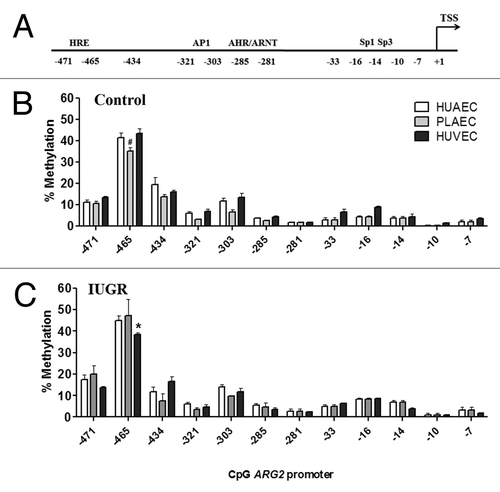
Figure 6. Comparison of DNA methylation status in the ARG2 promoter of normal and IUGR cells. Specific CpG methylation (%) in normal and IUGR-HUAEC (A), PLAEC (B) and HUVEC (C) determined by pyrosequencing of bisulfite treated DNA samples. Values are mean ± SEM ***p < 0.001, *p < 0.05, vs. corresponding control EC, two-way ANOVA.
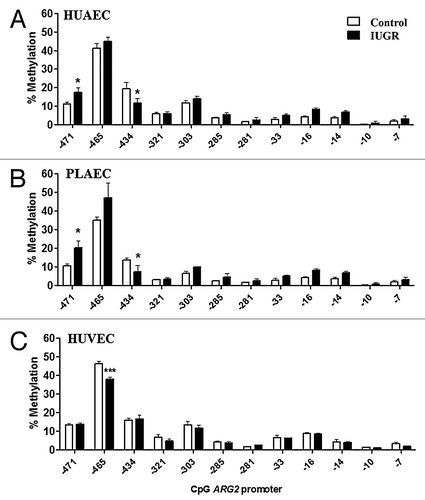
Figure 7. Effect of DNMT1 silencing on eNOS and arginase-2 mRNA levels in IUGR endothelial cells. Silencing of DNA methyltransferase-1 (DNMT-1) induced a concentration-dependent downregulation of this protein in HUAEC (A) and HUVEC (B). Quantification of eNOS and arginase-2 mRNA levels in IUGR-HUAEC (C and E, respectively) and IUGR-HUVEC (D and F, respectively) (solid bars, n = 5) in basal conditions (control), and cells treated with an non-specific siRNA (siScr) or specific siRNA against DNMT1 (siDNMT1). Levels of mRNA are expressed as 2-ΔΔCT referred to control condition and compared with basal levels in normal cells (open bars, n = 5). Values are mean ± SEM *p < 0.05, vs. control, one-way ANOVA.
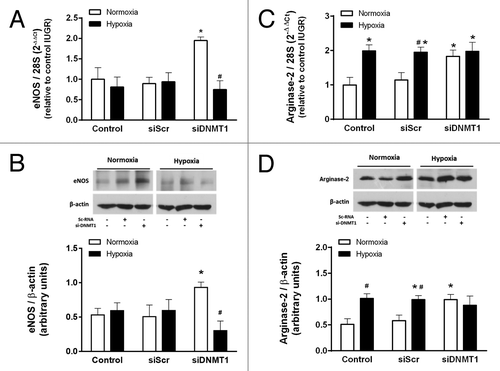
Figure 8. Effect of DNMT1 silencing on the response to hypoxia in IUGR-HUVEC. Quantification of eNOS (mRNA, A; protein, B) and arginase-2 (mRNA, C; protein, D) levels in IUGR-HUVEC exposed to normoxia (5% oxygen, open bars, n = 5) or hypoxia (2% oxygen, solid bars, n = 5) in basal conditions (control), or in the presence of an unspecific siRNA (siScr) and a siRNA against DNA methyltransferase-1 (siDNMT1). mRNA levels are expressed as 2-ΔΔCT referred to control condition in normoxia. Values are mean ± SEM *p < 0.05 vs. control, #p < 0.05 vs. corresponding condition in normoxia, two-way ANOVA.