Figures & data
Figure 1. ChIP-Seq analysis of H3K4me3 and H3K27me3 in uninduced (mK3, yellow) and induced (mK4, blue) metanephric mesenchyme cells.(A and B) Genomatix analysis of the relative distance of H3K4me3 and H3K27me3 active regions (peaks) to the transcription start sites (TSS) of actively transcribed genes in a 20 Kb window. (C and D) Top 5 GO Biological terms determined by GREAT analysis of categorized H3K4me and H3K27me3 active regions (yellow, mK3 unique; blue, mK4; red, shared).
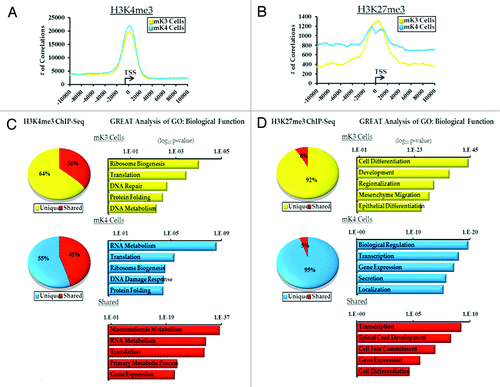
Figure 2. Chromatin platform of nephron progenitor renewal genes. Snapshots of H3K4me3 (green) and H3K27me3 (red) ChIP-Seq tracks of the progenitor genes Osr1 (A) and Six2 (B) in uninduced (mK3) and induced (mK4) cells. Differentiation is marked by loss of promoter H3K4me3 occupancy.
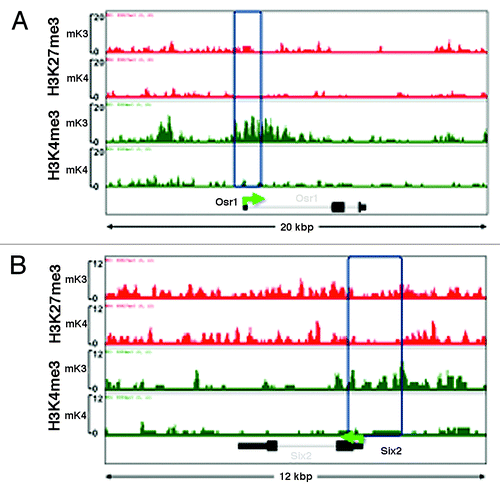
Figure 3.Six2 silencing in induced mK4 cells correlates with acquisition of a repressive chromatin signature. Six2 is expressed in uninduced mK3 but not induced mK4 cells. On the left side, low and high power snapshots of ChIP-Seq H3K4me3 (green) and H3K27me3 (red) tracks are shown. The right panel depicts ChIP-qPCR in the yellow-boxed region. Silencing of Six2 in mK4 cells is associated with loss of H3K4me3, gain of H3K4 demethylase, Kdm5b, and gain of H3K9me2 and methyltrasnferase G9a. Fold occupancy normalized to input and isotype-specific IgG controls. mK3 ChIP value is given a value of 1. n = 3 ChIP experiments per antibody. * p < 0.05 mK4 vs. mK3.
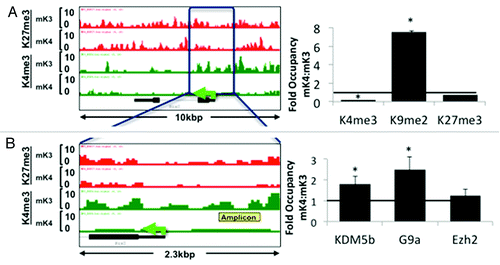
Figure 4. Chromatin platform of nephrogenic genes. Snapshots of H3K4me3 (green) and H3K27me3 (red) ChIP-Seq tracks of genes activated during differentiation. Two major chromatin patterns emerge during differentiation-associated gene activation: (1) loss of repressive H3K27me3 and gain of active H3K4me3 (A,C, and D); or (2) predominant gain of H3K4me3 (B,E, and F).
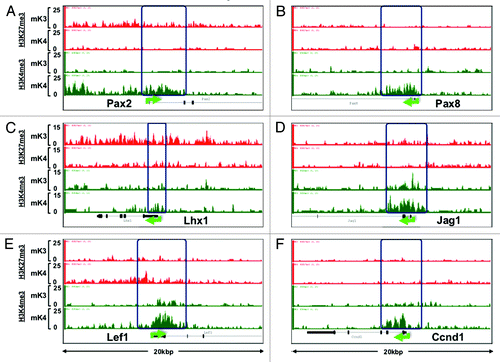
Figure 5. Nephrogenic gene expression correlates with acquisition of active chromatin signatures. On the left side of each panel, low (upper) and high (bottom) power snapshots of ChIP-Seq tracks. The right side of each panel depicts ChIP-qPCR of H3K4me3, H3K9me2 and H3K27me3 and respective modifiers around the yellow-boxed region. In each case, gene activation in mK4 cells is marked by gain of H3K4me3, loss of H3K9me2 and/or H3K27me3 and respective methyltransferases. Fold occupancy normalized to input and isotype-specific IgG controls. ChIP-PCR in mK3 was assigned a value of 1. * p < 0.05 mK4 vs. mK3.
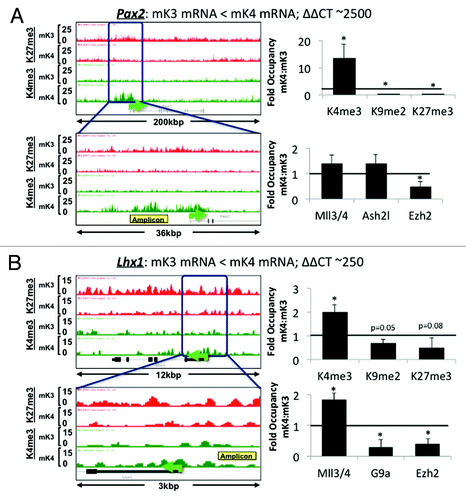
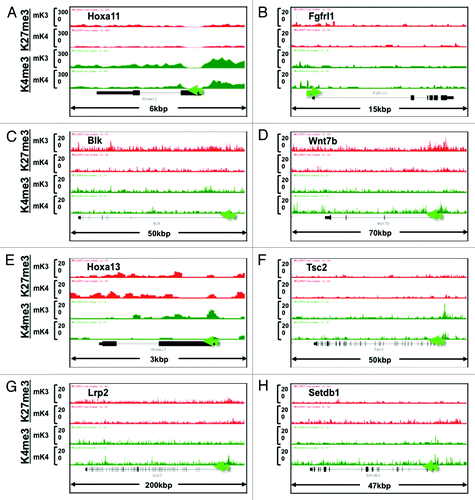
Figure 6. Chromatin signature of novel developmental genes expressed in mK3 or mK4 cells. (A–D) Genes expressed in mK3 cells are coated with the active histone mark H3K4me3 around the transcription start site. H3K4me3 peaks are absent when these genes are silent in mK4 cells. (E–H) Genes expressed in mK4 cells are coated with H3K4me3 peaks around the transcription start site. H3K4me3 peaks are absent when these genes are silent in mK3 cells. In the case of Myb (F), there is a net loss of H3K27me3 in mK4 vs. mK3.
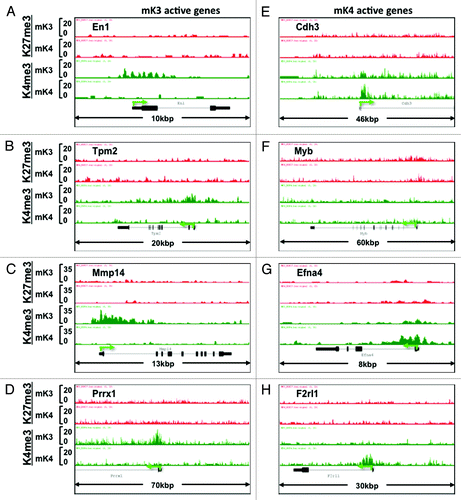
Figure 7. Chromatin signature of candidate developmental genes involved in human CAKUT and in regulation of renal function (see text for details). Transition from mK3 to mK4 cells correlates with gene expression and is associated with several patterns including retention of H3K4me3, loss of H3K27me3/gain of H3K4me3, or gain of H3K4me3 peaks. Loss of H3K4me3 (E and F) correlates with gene repression.