Figures & data
Figure 1. Flowchart of normalizations implemented. Ten color channel normalization procedures were implemented. Nine of those procedures were reference normalization factor (RN-factor) based methods that use the n = 93 normalization control probes assayed in every sample on the 450K chip for adjustment. Of the RN-factor based methods, three methods used the RN-factors from a single sample: the Illumina first sample normalization (IFSN), the best performing sample normalization, and the worst performing sample normalization. The remaining six RN-factor based procedures use aggregated RN-factors across different groups of samples, including the mean RN-factors for each plate of the experiment (Plates1–5 Means) and the all sample mean normalization (ASMN) that uses the mean RN-factors for all experimental samples. The remaining normalization, the lumi procedure, uses a quantile-based methodology instead of RN-factors.
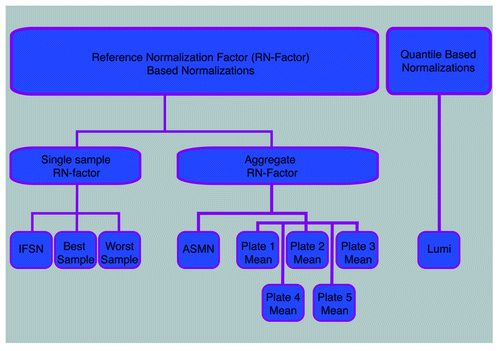
Figure 2. Reference normalization factor (RN-factor) based color channel normalization for the 450K methylation array. (A) The 450K chip includes n = 93 normalization control probes in both assay colors (red and green). The mean values of these sites are used to create RN-factors for normalizing both color channels over all samples (i.e., an experiment). The Illumina first sample normalization (IFSN) method uses the first sample’s mean red and green control probes as RN-factors ( and
). The all sample mean normalization (ASMN) method instead uses the mean read and green control probes taken across all control sites and all samples in a given experiment (
and
) as RN-factors. (B) A set of sample-wise normalization values, taken as the ratio of the RN-factor to each sample’s mean control probe values, is then computed. This results in a vector of length n normalization values for each color channel (R-RNV and G-RNV). (C) Color channel normalization of sample data occurs by multiplying the each of the jth sample’s red and green signals by the jth normalization value from the corresponding RN-vector (where j = 1,2,…, n).
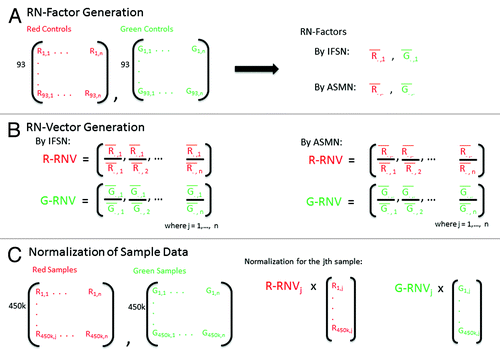
Figure 3. Plot of mean red (A) and green (B) signal intensity of normalization control probes (n = 93) by number of detected CpG sites in the 450K array sample data (n = 432). For both color channels, samples with lower intensity readings in their normalization control probes tended to have more poor performing CpG sites in their samples.
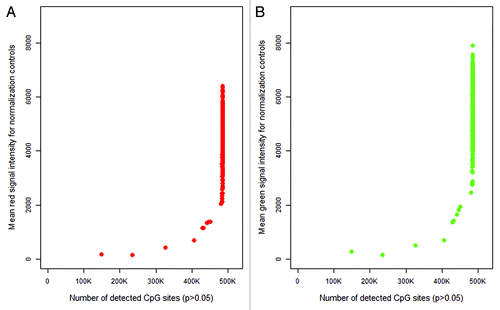
Figure 4. Mean control probe color signal intensity before and after normalization. (A) Distribution of mean green and red normalization controls (93 controls per signal color per sample) as included in the 450K chip over 432 DNA samples. Each point, red triangle or green square, represents the average of the normalization controls for that signal color per sample prior to implementation of color channel normalization. (B) Following adjustment using a reference normalization factor (RN-factor) based normalization, the average normalization controls for all samples are ‘forced’ to be the same level, making observations across samples comparable. Here, ASMN normalization was performed which uses the mean red and green signal for all samples for adjustment.
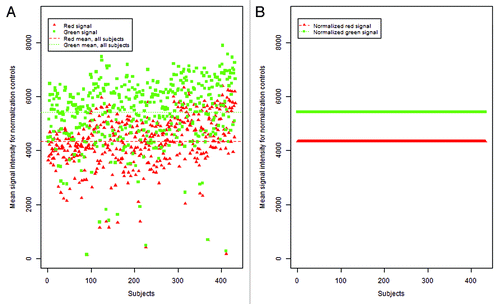
Figure 5. Plot of normalized DNA methylation (βs) given an unadjusted β of 0.1 (Signal A = 5000 and Signal B = 570) for all 432 samples. Open circles represent data normalized using the sample with the least detectable sites (sample 411, the lowest quality sample). Filled circles were normalized using the sample with the most detectable sites (sample 355, the highest quality sample).
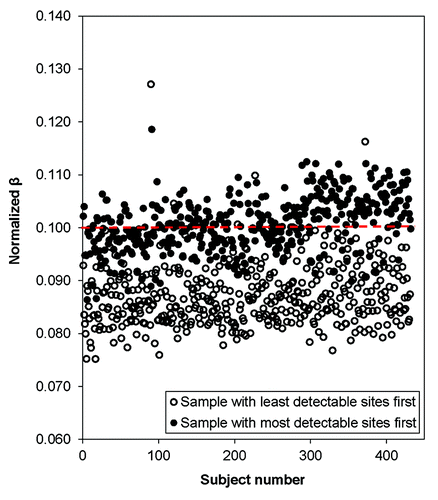
Figure 6. Average percent change of methylation values, βs, after normalization by best and worth performing samples. Mean percent change in βs, values ranging from 0.1 to 0.9, based on normalization by the lowest quality sample (largest amount of CpG sites with p < 0.05) and the highest quality sample (least amount of CpG sites with p < 0.05) over all samples (n = 432). While normalization by the highest quality sample changed the βs only slightly (< 10% on average), normalization by the lowest quality sample tended to change the low and high methylation βs substantially (> 10% on average).
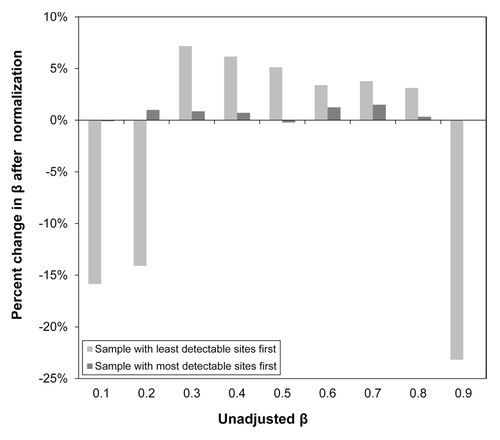
Table 1. Reference normalization factors and methylation (βs) for a single sample by normalization procedure
Table 2. Repeatability of technical replicates by improvement of root mean squared error (root-MSE) and mean Spearman correlation (R2) compared for un-normalized results
Figure 7. Box plots of sample mean methylation by normalization methods. Box plots of mean per-sample methylation (β) for all sites interrogated on the 450K array (n = 485 512) by color channel normalization methods. Plots are shown for (A) un-normalized results and three different normalization methods, (B) lumi smooth quantile normalization, (C) normalization using the worst performing sample’s reference normalization factor values (sample 411), and (D) using the all sample mean normalization (ASMN) method. Each chip assays 12 samples, so every box plot contains 12 observations in total.
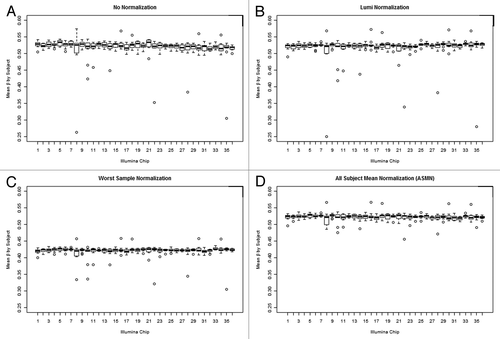
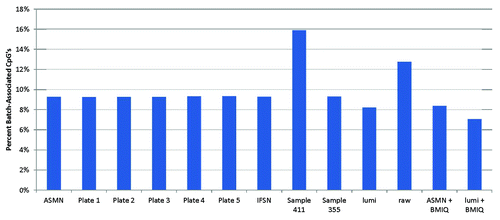
Figure 8. Percent of 450K array CpG sites associated with chip batch (p < 0.01) shown by normalization method. Normalization methods include: All sample mean normalization (ASMN), normalization by reference normalization factors (RN-factors) taken as the mean control probe values for each of the plates (1–5) run, Illumina first sample normalization (IFSN), normalization by the worst performing sample’s RN-factors (sample 411) and the best performing sample’s RN-factors (sample 355), lumi smooth quantile normalization, raw un-normalized results, and both the ASMN and lumi normalization followed by β-mixture quantile normalization (BMIQ). Batch association was evaluated by ANOVA for each of the n = 485 512 CpG sites interrogated.