Figures & data
Figure 1. KDM5B and KDM5C are covalently modified by SUMO. (A) SUMOylation of KDM5B and KDM5C, but not KDM5D. Whole cell lysates from HEK293T cells transfected with FLAG-KDM5B, MYC-KDM5C or FLAG-KDM5D in combination with HA-UBC9/HIS-SUMO-1 or HA-UBC9/HIS-ΔGGSUMO-1 were immunoblotted with the indicated antibodies. The molecular mass markers are shown on the left in kDa and the migration of the unmodified and modified KDM5 family members is indicated on the right. Asterisks (*) indicate high molecular weight SUMOylated proteins in the lower panels. (B) KDM5B is SUMOylated by SUMO-1 and SUMO-3. Whole cell lysates from HEK293T cells transfected with FLAG-KDM5B alone or in combination with HIS-SUMO-1 or HIS-SUMO-3 in the presence or absence of HA-UBC9 were immunoblotted with the indicated antibodies. The molecular mass markers are shown on the left in kDa and the migration of the unmodified and modified KDM5B is indicated on the right. Asterisk indicates high molecular weight proteins conjugated by HIS-tagged SUMO-1 and SUMO-3. (C) SUMO-3 is covalently conjugated to KDM5B. HIS-SUMO-3 conjugated proteins were purified under denaturing conditions from total cell lysates of HEK293T cells expressing FLAG-KDM5B alone or in combination with HIS-SUMO-3. SUMOylated proteins bound to Ni-NTA agarose beads were eluted and immunoblotted with anti-FLAG antibodies. The migration of SUMO-3-FLAG-KDM5B is shown on the right. (D) Endogenous KDM5B is SUMOylated. Total cell lysates from MCF-7 cells stably expressing HIS-SUMO-3 were incubated with Ni-NTA agarose beads under denaturing conditions. SUMOylated proteins bound to beads were then immunoblotted with a polyclonal antibody against KDM5B. An equal number of MCF7 cells lacking expression of HIS-SUMO-3 were used as a control. The migration of endogenous SUMO-3-KDM5B is shown on the right and the arrowhead indicates a non-specific band.
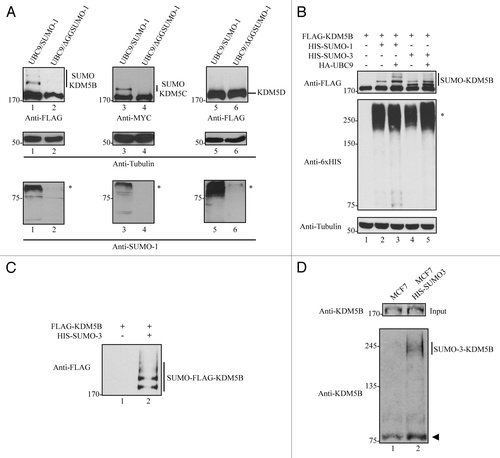
Figure 2. hPC2 enhances the SUMOylation of KDM5B. (A) KDM5B co-localizes with hPC2 in U2OS cells. DAPI-stained nuclei (blue) and ectopically expressed FLAG-KDM5B (red), and YFP-hPC2 (green) were detected by fluorescence microscopy. The scale bar denotes 5 µm and images were merged in the indicated panel. Arrowheads indicate transfected cells. (B) SUMOylation of KDM5B is enhanced by hPC2 and not PIASy. Whole cell extracts from HEK293T cells expressing FLAG-KDM5B and HIS-SUMO-3 alone or in combination with YFP-hPC2 or Myc-tagged PIASy were subjected to immunoblotting with the indicated antibodies on the left. The migration of SUMO-3-FLAG-KDM5B is shown on the right. The migration of protein molecular weight markers is shown on the left in kDa.
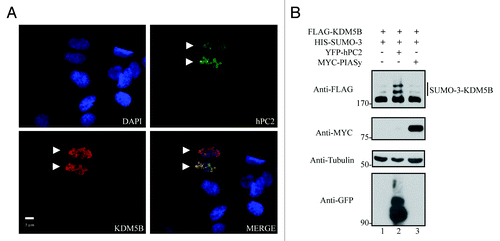
Figure 3. Mapping of the SUMO-acceptor sites of KDM5B. (A) Diagram depicting the distribution of the protein domains found in human KDM5B and the localization of two putative SUMOylation sites, K242, and K278. Clustal W amino acid sequence alignment of KDM5B from human (Homo sapiens; NP_006609), mouse (Mus musculus; NP_690855), chicken (Gallus gallus; NP_001026200), frog (Xenopus tropicalis; XP_002939684), and fish (Danio rerio; Q6IQX0) is depicted. The evolutionarily conserved putative SUMO-acceptor sites K242 and K278 are represented in bold. The abbreviations are JmjN (Jumonji N) domain; ARID (AT-rich interaction domain); PHD (plant homeobox domain), JmjC (Jumonji C) domain; and C5HC2 (Zinc finger domain). (B) KDM5B is SUMOylated by SUMO-3 at residues K242 and K278. Whole cell lysates obtained from HEK293T cells expressing HIS-SUMO-3, YFP-hPC2 and FLAG-KDM5BWT (wild type) or mutants K242R, K278R, and 2KR (K242R and K278R) were immunoblotted with the indicated antibodies. Asterisks (*) and (**) indicate high molecular weight SUMOylated proteins and non-conjugated SUMO-3, respectively.
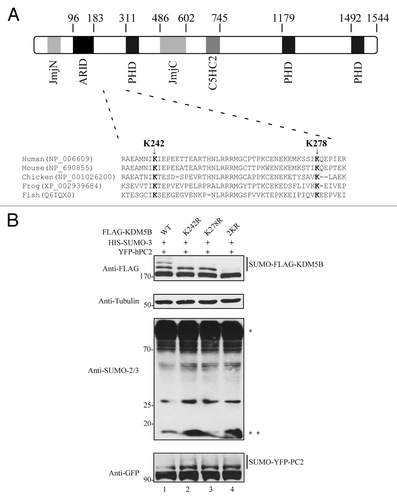
Figure 4. SUMOylated and unSUMOylated KDM5B protein levels are regulated along the cell cycle. (A) KDM5B protein levels are upregulated in mitotic cells. MEF and U2OS cells were arrested at the G2/M phase with nocodazole for 22 h or not (asynchronous) and total cell lysates subjected to immunoblotting with anti-KDM5B and anti-Tubulin (loading control) antibodies. Relative KDM5B protein amounts (bottom of KDM5B immunoblots) were calculated based on band intensities quantified using ImageJCitation45 and normalized to Tubulin levels. (B) HEK293T cells ectopically expressing FLAG-KDM5B were arrested or not (asynchronous) at the G2/M and G1/S phase following 22 h of treatment with nocodazole or hydroxyurea, respectively. Whole cell lysates were analyzed by immunoblotting with anti-FLAG and anti-Tubulin (loading control) antibodies. Relative KDM5B protein amounts (bottom of FLAG immunoblot) were calculated based on band intensities quantified using ImageJCitation45 and normalized with Tubulin. (C) Levels of SUMOylated KDM5B are increased in cells arrested at the G2/M and G1/S phases. HEK293T cells were transfected with expression vectors encoding FLAG-KDM5B, HIS-SUMO-3, and YFP-hPC2. Cells were arrested or not (asynchronous) at the G2/M (nocodazole) and G1/S (hydroxyurea) phase. Immunoblots were performed using total cell lysates and antibodies against FLAG and Tubulin (loading control). (D) SUMOylation of KDM5B is regulated during the cell cycle. HEK293T cells expressing KDM5B, SUMO-3, and hPC2 were arrested at the G1/S phase by treatment with hydroxyurea for 22 h. Subsequently, cells were released in fresh culture medium to allow transition to G2/M. Whole cell lysates were then collected every 2 h for a period of 8 h. Levels of SUMOylated and unSUMOylated KDM5B were detected by immunoblotting with anti-FLAG antibodies. Cell cycle progression was monitored by immunoblotting with anti-Cyclin B1 antibodies. The ratio between the amounts of SUMOylated vs. unSUMOylated KDM5B (bottom of FLAG immunoblot) was calculated based on band intensities quantified using ImageJCitation45 and normalized with Tubulin. (E) RNF4 promotes degradation of KDM5B. HEK293T cells were transfected with FLAG-KDM5B and HIS-SUMO-3 plasmids in the absence or presence of increasing amounts of an expression vector encoding FLAG-RNF4. Total cell lysates were immunoblotted with anti-FLAG antibodies to detect KDM5B or RNF4, as indicated. The migration of FLAG-KDM5B and FLAG-RNF4 is shown on the right. The antibodies used are indicated on the left with the migration of the molecular mass markers in kDa. (F) Degradation of KDM5B by RNF-4 is mediated by the proteasomal pathway. HEK293T cells expressing FLAG-KDM5B in the presence or absence of FLAG-RNF4 were treated or not with MG132 for 6 h. Total cell lysates were immunoblotted with the antibodies indicated on the left. The migration of FLAG-KDM5B and FLAG-RNF4 is shown on the right. The antibodies used are indicated on the left with the migration of the molecular mass markers in kDa.
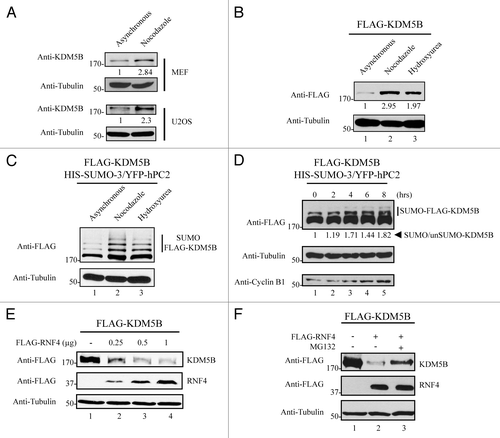
Figure 5. SUMOylation modulates the occupancy of KDM5B at target genes. (A) ChIP assays were performed using HEK293T transfected with vectors encoding HIS-SUMO-3, YFP-hPC2, and KDM5BWT-eGFP or KDM5B2KR-eGFP. Equivalent expression of KDM5B-eGFP proteins was examined by immunoblotting total cell lysates with the indicated antibodies. Slower migrating forms representing SUMOylated KDM5B were more abundant in the WT vs. the 2KR mutant. (B) HEK293T cells expressing SUMO-3, hPC2, and KDM5BWT-eGFP or KDM5B2KR-eGFP were subjected to chromatin immunoprecipitations with antibodies against GFP or control IgG. The occupancy of KDM5BWT-eGFP or KDM5B2KR-eGFP at the promoter regions of the genes STARD4, miR-1792, RB1CC1, and BMI-1 was evaluated by real-time qPCR. Error bars represent ± 1 SD of triplicate qPCR reactions. The asterisk (*) indicates p < 0.05 (Student’s t-test). (C) ChIP assays were performed with antibodies against histone 3 (H3), H3K4me3, and immunoglobulin G using HEK293T cells expressing SUMO-3, hPC2, and KDM5BWT-eGFP or KDM5B2KR-eGFP. The ChIP assays were normalized to H3 levels. Relative levels of trimethylated H3K4 present at the promoter regions of the genes STARD4, miR-1792, RB1CC1, and BMI-1 were evaluated by real-time qPCR. Error bars represent ± 1 SD of triplicate qPCR reactions. The asterisk (*) indicates p < 0.05 (Student’s t-test). (D) HEK293T cells were co-transfected with vectors encoding HIS-SUMO-3 and FLAG-hPC2 (Control) or in combination with plasmids expressing KDM5BWT-eGFP (WT) or KDM5B2KR-eGFP (2KR). Total RNA was isolated, reverse transcribed and mRNA expression levels of STARD4, RB1CC1 and BMI-1 were evaluated by qPCR. Results were normalized according to the expression levels of the house keeping genes GAPDH and CLTC. Error bars represent ± 1 SD of triplicate qPCR reactions. The asterisk (*) indicates p < 0.05 (Student’s t-test).
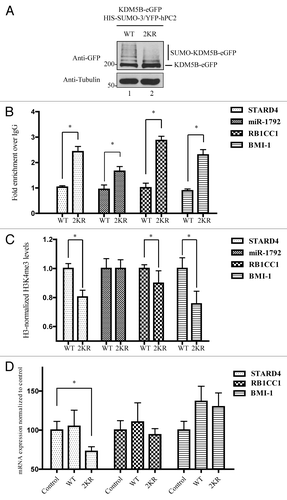
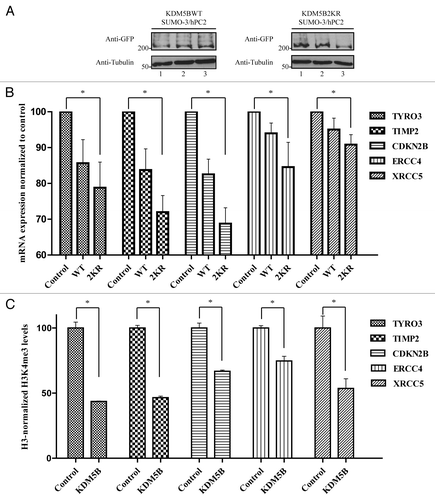
Figure 6. SUMOylation-deficient KDM5B regulates the expression of a unique set of genes. (A) HEK293T cells were co-transfected with vectors encoding HIS-SUMO-3 and FLAG-hPC2 in combination with plasmids expressing KDM5BWT-eGFP or KDM5B2KR-eGFP. Immunoblotting of total cell lysates with anti-GFP and anti-Tubulin antibodies revealed similar levels of KDM5BWT-eGFP and KDM5B2KR-eGFP across the triplicate transfections used for the digital gene expression analysis. (B) Total RNA isolated from biological triplicates of HEK293T cells transfected with vectors encoding for HIS-SUMO-3 and FLAG-hPC2 (Control) or in combination with plasmids expressing KDM5BWT-eGFP (WT) or KDM5B2KR-eGFP (2KR) was subjected to digital gene expression analysis (NanoString). The bar graph depicts the mRNA expression levels of 5 out of 12 genes found to be significantly downregulated only in cells expressing KDM5B2KR-eGFP. The asterisk (*) indicates p < 0.05 (Student’s t-test). (C) Levels of H3K4me3 at the promoter region of the genes illustrated in panel B were assessed in mock (Control) or KDM5B-eGFP-transfected HEK293T cells. ChIP assays were performed with antibodies against histone 3 (H3), H3K4me3, and immunoglobulin G. The asterisk (*) indicates p < 0.05 (the Student t-test).