Figures & data
Figure 1. Strategy for site-specific DNA methylation. Step1, Conventional reporter constructs are adapted as template. A pair of primers (arrows) containing synthetically methylated CpGs (stars) are phosphorylated at 5′ end. Both primers are extended by high-fidelity DNA polymerase to generate new complementary strand DNA containing a nick. The new complementary strand DNA is ligated to form circular DNA by Taq DNA ligase. Step 2, The parental DNA template is digested by Dpn1 which recognizes 5′-Gm6ATC-3′, which are initially derived from E. coli., and the resulting linear DNA is digested by T7 exonuclease. Step 3, Dnmt1 is used to methylate the remaining DNA strand complementary to the methylated primer. Step 4, The integrity of the intended methylated CpG sites is confirmed by bisulfite sequencing before transfection.
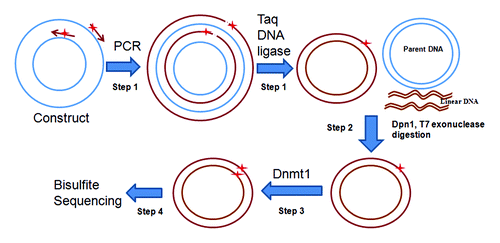
Figure 2. PCR-ligase coupled amplification of DAPK reporter constructs. Two primers containing four methylated CpGs were used for PCR-ligase coupled amplification of DAPK reporter constructs (5011bp). (A) pfuUltra II Fusion HS DNA polymerase (lane 1) and Herculase II Fusion DNA polymerase (lane 2) could amplify the constructs along with Taq DNA ligase. (B) PCR-ligation products were digested with Dpn1 and T7 exonuclease, lane 1: pfuUltra II Fusion HS DNA polymerase product; lane 2: Herculase II Fusion DNA polymerase product; lane 3: 1kb plus DNA ladder.
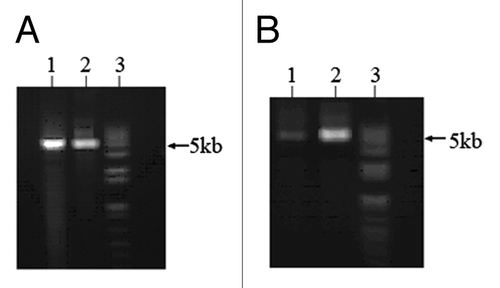
Figure 3. Bisulfite sequencing of the DAPK promoter in site-specific methylated reporter constructs. Site-specific methylated constructs (MP2) were used for bisulfite treatment. The promoter region was amplified with vector specific primers flanking the promoter. PCR product was subject to direct Sanger sequencing.Citation25 (A) The sequencing result before transfection into A549 cells. (B) The sequencing result after transfection into A549 cells for 36 h. (C) The anti-sense strand sequencing result after transfection into A549 cells for 36 h. The upper sequence in each pair is genome sequence in which the underlined target sequence containing four methylated cytosines are in red characters. In the bisulfite sequencing, all unmethylated cytosines were converted to thymine. Only the four intended target cytosines were methylated in the bisulfite sequencing result. Corresponding chromatograms are depicted in the supplement (Fig. S1).
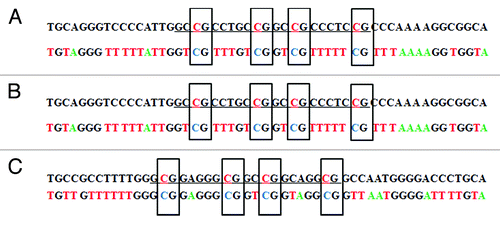
Figure 4. Functional analysis of CpG methylation impact on DAPK promoter activity. Regions ~60 bp in length (30 bp forward and 30 bp reverse primers fit end-to-end) were tested. The numbers of methylated CpG sites were determined by the endogenous sequence in that 60 bp region (e.g., four CpGs at -10, -23, and -34 for MP1, three CpGs for MP2, five CpGs for MP3, seven CpGs for MP4, nine CpGs for MP5 and three CpGs for MP6). Unmethylated DAPK reporter constructs (unM) and six site-specific methylated DAPK reporter constructs (MP1 to MP6) were transfected into A549 cells, PRL-TK which expresses Renilla luciferase was co-transfected as an internal control. The empty vector (Basic) was also transfected as negative control. All constructs were transfected in triplicates. After 36 h, luciferase activities were determined by dual luciferase assay. Firefly luciferase activity was normalized to Renilla luciferase activity (internal control). Here, the most 3′ cluster (the three CpGs methylated together by the MP1 patch, which covers the region -34 to -10, reference to TSS) appears to halve the promoter activity. P values represent t test comparisons with the unmethylated DAPK promoter insert (unM) control. KEY: Basic: the reporter construct without promoter; unM: The reporter construct with unmethylated DAPK promoter; MP1 to MP6: reporter constructs with DAPK promoter methylated at different CpG sites; mCpG: Methylated CpG.
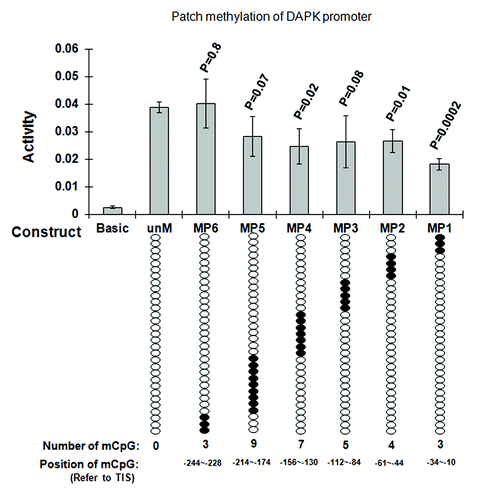
Table 1. Proteins differentially binding to methylated or unmethylated DNA oligos repeating DAPK and RASSF1A proximal promoters by LC-MS/MS
Figure 5. Functional analysis of DNA methylation impact on RASSF1A promoter reporter constructs. Unmethylated RASSF1A reporter constructs (unM) and four site-specific methylated reporter constructs (MP1 to MP4) were transfected into A549 cells. PRL-TK, which expresses Renilla luciferase was co-transfected as an internal control. The empty vector (Basic) was also transfected as negative control. All constructs were transfected in triplicates. After 36 h, luciferase activities were determined by dual luciferase assay. Firefly luciferase activity was normalized to Renilla luciferase activity (internal control). Here, each of two clusters, each containing four methylated CpGs at +21,+15,+2,-6 and -138, -129,-125,-102, reference to TSS, appear to halve promoter activity. P values represent t test comparisons with the unmethylated RASSF1A promoter insert (unM) control. Basic, the reporter construct without promoter; unM, the reporter construct with unmethylated RASSF1A promoter; MP1 to MP4, reporter constructs with RASSF1A promoter methylated at different CpG sites; mCpG, methylated CpG.
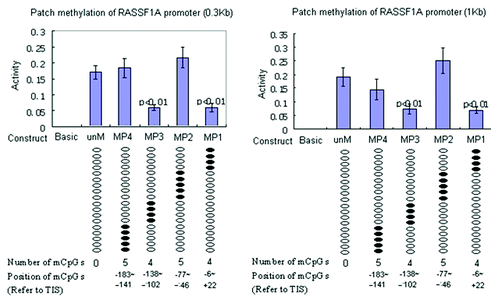
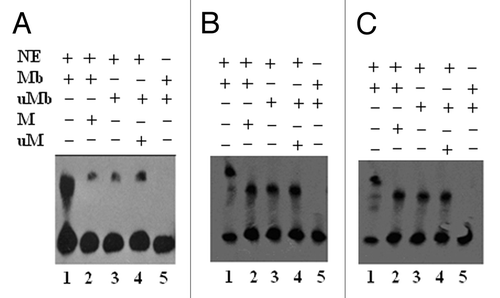
Figure 6. LightShift EMSA for biotin labeled methylated and unmethylated DAPK-MP1, RASSF1A –MP1, and MP3 oligos from the DAPK and RASSF1A promoter. (A): Lane 1: Nuclear extract proteins specifically bind to the methylated DAPK-MP1 probe. Lane 2: The binding of nuclear extract proteins to the methylated DAPK-MP1 was competed with unlabeled methylated DAPK-MP1. Lane 3: Nuclear extract proteins nonspecifically bind to unmethylated DAPK-P1 probe. Lane 4: The nonspecific binding of nuclear extract proteins to the unmethylated DAPK-P1 was not competed with unlabeled unmethylated DAPK-P1. Lane 5: Methylated DAPK-MP1 only as negative control. (B): Lane 1: Nuclear extract proteins specifically bind to the methylated RASSF1A-MP1. Lane 2: The binding of nuclear extract proteins to the methylated RASSF1A-MP1 was partially inhibited by unlabeled methylated RASSF1A-MP1. Lane 3: Nuclear extract proteins bind to unmethylated RASSF1A-P1. Lane 4: The binding of nuclear extract proteins to the unmethylated RASSF1A-P1 was competed with unlabeled unmethylated RASSF1A-P1. Lane 5: RASSF1A-MP1 only as negative control. (C): Lane 1: Nuclear extract proteins specifically bind to the methylated RASSF1A-MP3. Lane 2: The binding of nuclear extract proteins to the methylated RASSF1A-MP3 was partially inhibited by unlabeled methylated RASSF1A-MP3. Lane 3: Nuclear extract proteins bind to unmethylated RASSF1A-P3. Lane 4: The binding of nuclear extract proteins to the unmethylated RASSF1A-P3 was competed with unlabeled unmethylated RASSF1A-P3. Lane 5: RASSF1A-MP3 only as negative control. NE: nuclear extracts; Mb: methylated DNA labeled with biotin; M: methylated DNA unlabeled; uMb: unmethylated DNA labeled with biotin; uM: unmethylated DNA unlabeled.