Figures & data
Figure 1. Technical validation of 450K array data by sodium bisulfite pyrosequencing. Correlation of DNA methylation as measured by array β-value and by bisulfite pyrosequencing for 70 individual sites in T-lymphocytes and B-lymphocytes from seven of the ten healthy individuals (5 CpGs were selected at random, from three separate genes: HMOX2: cg14951292, AMN: cg09616556, PM20D1: cg24503407, cg07167872, cg11965913). T- and B-lymphocytes are shown as circles and crosses, respectively. Spearman’s r = 0.952, P < 0.000 01.
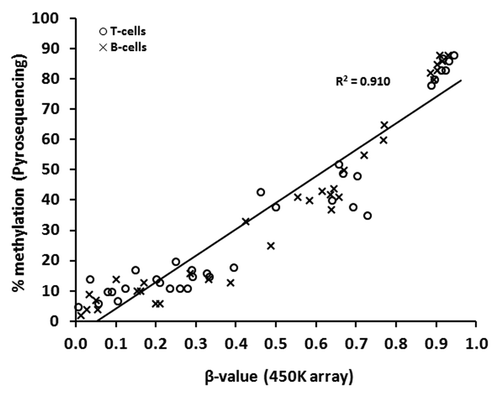
Figure 2. LINE-1 DNA methylation in matched pairs of T-lymphocytes and B-lymphocytes. Sodium bisulfite pyrosequencing was used to quantify methylation at three CpG sites within LINE-1 repetitive sequences in purified T-lymphocytes and B-lymphocytes from ten healthy individuals. T- and B-lymphocytes are shown as circles and crosses, respectively. The mean level of methylation for each CpG site in each cell population is shown by the short black horizontal bar in each case. * P ≤ 0.01 (Wilcoxon Signed-Rank Test).
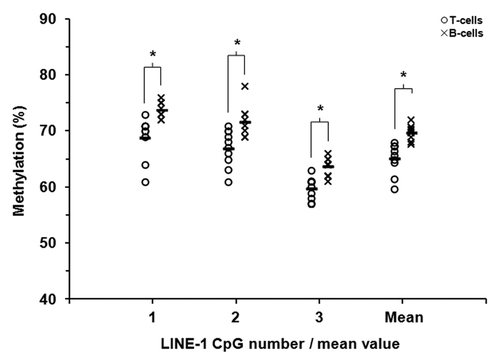
Figure 3. Criteria for filtering and identification of CpGs differentially methylated between paired T-lymphocyte and B-lymphocyte samples. The starting number of 484,616 CpGs was derived through removal of CpGs with high detection P values (P > 0.05) and those with missing β-values, as described in the Methods. †, “change” refers to a methylation difference that fulfilled all preceding filtering steps; ‡, “change in the same direction” refers to multiple CpGs from one gene all showing either hyper- or hypo-methylation in one cell type as compared with the other cell type.
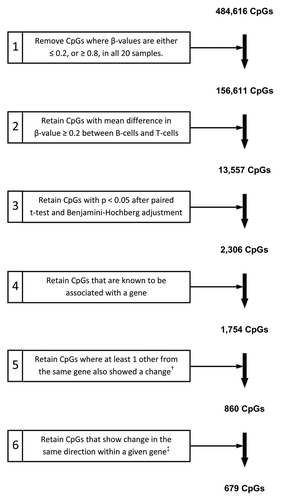
Figure 4. Heatmap and clustering for the 679 CpGs identified as differentially methylated. CpG sites identified by 450K array analysis as differentially methylated between paired T-lymphocytes (green bar) and B-lymphocytes (red bar) were analyzed by hierarchical clustering. Each row represents an individual CpG site and each column a different sample (listed beneath the heatmap). Branches of the dendrograms indicate similarity between CpGs (rows) and samples (columns). Color gradation from yellow to blue represents low to high DNA methylation respectively, with β-values ranging from 0 (no methylation; yellow) to 1 (complete methylation; blue). Sample order, left-to-right: B-lymphocytes: HC03, 10, 04, 09, 11, 14, 08, 12, 16, 17; and T-lymphocytes: HC03, 08, 09, 11, 12, 04, 14, 16, 10, 17.
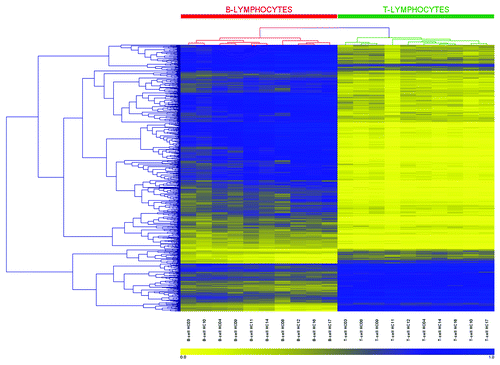
Table 1. Summary characteristics of the 679 CpGs identified as differentially methylated between paired T-lymphocyte and B-lymphocyte samples by genome-wide DNA methylation analysis using 450K arrays
Table 2. Annotation for the top 10 hypermethylated genes in T-lymphocytes and top 10 hypermethylated genes in B-lymphocytes, as determined by genome-wide DNA methylation analysis
Figure 5. Validation of differentially methylated CpG candidates identified by 450K array analysis. Sodium bisulfite pyrosequencing was used to confirm candidate genes/CpGs that were differentially methylated between T-lymphocytes and B-lymphocytes in all ten healthy individuals. CpG sites in four genes that were hypermethylated in T-lymphocytes (A) and four genes that were hypermethylated in B-lymphocytes (B) were selected from the array candidates. T- and B-lymphocytes are shown by circles and crosses, respectively. Gene names are shown on the x-axis, where for each gene methylation values are shown for the array (left) and pyrosequencing (right). The mean values are defined by the horizontal black bar in each case. The methylation differences observed between T-lymphocytes and B-lymphocytes by pyrosequencing analysis were statistically significant for each of the eight CpGs/genes examined (P < 0.01, Wilcoxon Signed-Rank Test). Abbreviations: Pyro., bisulfite pyrosequencing.
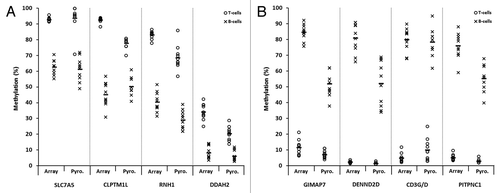
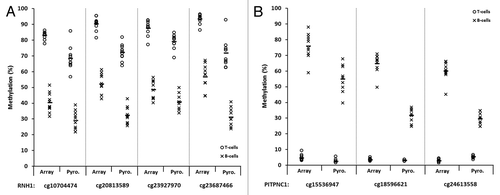
Figure 6. Representative plots displaying validation of multiple array CpGs within two candidate genes by sodium bisulfite pyrosequencing analysis. Four CpGs within the RNH1 gene (A) and three within PITPNC1 (B), all of which demonstrated significant differences in methylation between T-lymphocytes and B-lymphocytes by array analysis, were examined. T- and B-lymphocytes are shown as circles and crosses, respectively. CpG identifiers are shown on the x-axis. In each case, the first site shown for RNH1 (A) and PITPNC1 (B) is the site depicted in , respectively. For each site, methylation values are shown for the array (left) and pyrosequencing (right). The mean values are defined by the horizontal black bar in each case. The CpG sites are displayed in the 5′ to 3′ direction, with the four sites in RNH1 covering 20-bp and three sites in PITPNC1 covering 45-bp. The methylation differences observed between T-lymphocytes and B-lymphocytes by pyrosequencing analysis were statistically significant for each of the CpGs in each gene examined (P < 0.01, Wilcoxon Signed-Rank Test). Abbreviations: Pyro., bisulfite pyrosequencing.