Figures & data
Figure 1. (A) Four ZF-TDG constructs (‘TDG’) were transduced into 3T3 fibroblasts. Controls included 4 constructs with mutant catalytically inactive TDG (‘mut’), ZF-only (‘ZF’-) and mock-transduced cells (not shown). (B) To analyze methylation changes induced by the constructs, we performed pyrosequencing assay to positions in the promoter (top, 11:78734001–78734136) and in the nearest CpG island (bottom, 11:78738499–78738571). We found statistically significant decrease in methylation at most sites in the ZF-TDG group (TDG) vs. mutant control (mut) or ZF-only control (ZF). Interestingly, even the 10–20% reduction in methylation at these sites was associated with a strong transcriptional response (), which emphasizes the potential of DNA demethylation for manipulation of gene expression. Of note, not all CpG sites (6 out of 8 tested) showed decreased methylation suggesting some CpG are causative for transcription and some are not. (C) To compare the magnitude of the effect we interrogated these sites in cells transduced with single ZF-TDG constructs and respective ZF-alone controls. Decreases in methylation in these cells were substantially less prominent, as shown in these three representative sites. All experiments were performed at least 3 times in triplicates, pyrosequencing measurements were performed in quadruplicates, *P < 0.05.
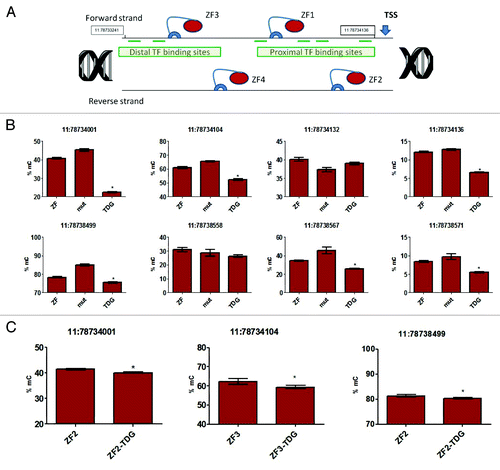
Figure 2. To confirm the binding of ZF constructs to desired DNA target areas in the promoter we performed ChIP-PCR using a standard sonication-based shearing protocol followed by Myc precipitation (ChIP kit, Sigma) and real-time PCR for a set of primers amplifying a small fragment of the Nos2 promoter. Both ZF2-TDG and the control ZF2 construct include Myc marker and, as evident, precipitated the desired fragment in Nos2 promoter indicating they were bound. ZF1-TDG uses FLAG instead of Myc, while ZF3-TDG is located ~700 bp away from ZF2 and is outside of the amplicon; they served as biological negative controls in the assay, together with control IgG they indicate that non-specific precipitation was minimal. Unprecipitated input DNA (diluted 1:100) served as positive control. *P < 0.05.
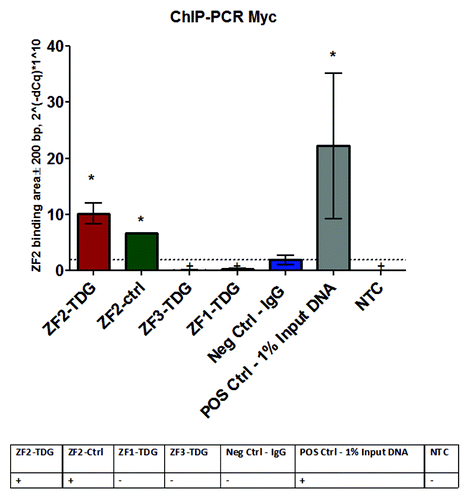
Figure 3. (A–D) Four simultaneous ZF-TDG constructs provided a strong transcriptional enhancement of Nos2 in response to stimuli without affecting background expression. Cells were stimulated either by LPS (0.5 ng/mL for 2 h) (A and B); or by IFNγ (200 U/mL 24 h) (C and D). Expression is normalized to β-actin. In B and D fold upregulation was obtained by dividing expression after stimulation to background unstimulated expression in that group. (E and F) To identify the efficiency of each construct separately, we transduced cells with each ZF-TDG or its ZF-only control (1+ vs. 1-, etc.) and tested effects on upregulation of Nos2 in identical conditions. ZF-TDG #2 led to the greatest increase in Nos2 transcription > #1 > > ##s 4 and 3. Interestingly, ZF #2 was the closest to TSS, followed by #1 and #4, with #3 being the most remote, which suggests that proximity to TSS may play a role. Notably, the fold change by each single construct was substantially lower than the cumulative effect of all 4, most prominently for the case of LPS, as seen in (G) where the fold upregulation is normalized to the level in Mock. All experiments were performed at least 3 times in triplicates, *P < 0.05; **P < 0.01.
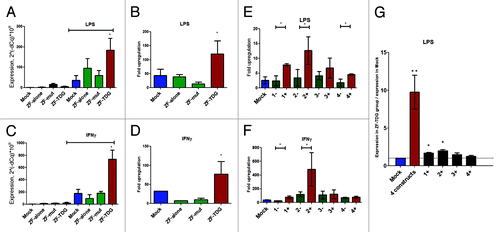
Figure 4. To determine gene-specificity of the effect microarray analysis was performed over LPS-stimulated and control RNA samples from cells carrying ZF-TDG and ZF-mutant control TDG. (A) RMA values were extracted using RMA Express; after quality control and outlier removal the resultant RMA matrix was analyzed in TIGR MeV software by factorial ANOVA to identify genes upregulated after LPS exposure only in the TDG, and not in the Mut groups with at least 1.1 fold. (B) Pathway analysis of the list in 4A using GeneGo MetaCore indicates that all except 4 of the genes are linked to Nos2 in a “shortest paths” network over < 2 intermediaries, indicating that differential elevation of these genes may be secondary to upregulation of Nos2.
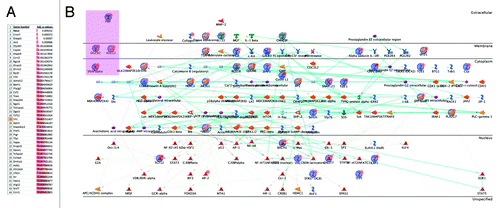
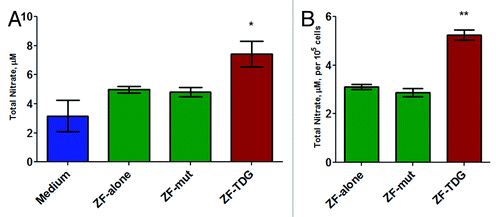
Figure 5. In cells expressing all 4 constructs, NO production was measured in the supernatants (phenol-free DMEM complete) after 48 h of 1 ng/mL LPS stimulation using the colorimetric Griess method. (A) The values are total nitrate after nitrite-to-nitrate conversion. (B) Total nitrate normalized to cell counts per well. All experiments were performed at least 3 times in triplicates, *P < 0.05; **P < 0.01.