Figures & data
Figure 1. Unsupervised clustering analysis of the normal cervical tissue (N), CIN3 tissue (CIN3), and the cervical cancer tissue (CC). The color gradient green to red displays the β-value and can range from 0–1.
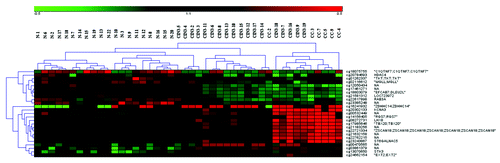
Table 1. Mean β-values for CpG sites represented in the heat map in
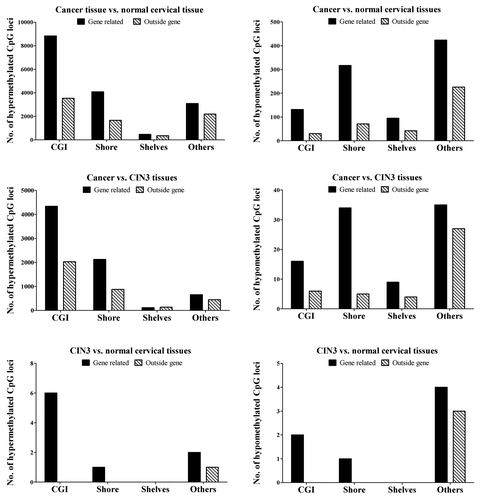
Figure 2. Distribution of hypermethylated and hypomethylated CpG locus in CpG island (CGI), CpG island shore, CpG island shelves and other regions.