Figures & data
Figure 1. Model illustrating the role of pRNA in recruiting chromatin modifying enzymes to rDNA. Transcripts that match the rDNA promoter, dubbed pRNA (promoter-associated RNA), form a specific secondary structure that is recognized by TIP5, the large subunit of the chromatin remodeling complex NoRC. NoRC is associated with histone deacetylases (HDACs) and histone methyltransferases (HMTs) that establish heterochromatic features at the rDNA promoter. In addition, pRNA directly interacts with DNA, forming a DNA:DNA:RNA triple helix with the binding site of the transcription factor TTF-I, leading to displacement of TTF-I. The triple helical structure is recognized by the DNA methyltransferase DNMT3b, which methylates the rDNA promoter, leading to transcriptional silencing.
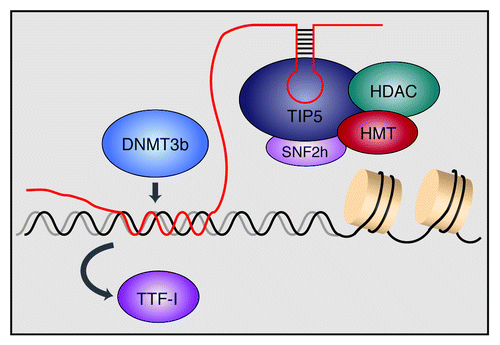
Figure 2. PAPAS induces trimethylation of H4K20 and chromatin compaction in growth-arrested cells. The scheme depicts epigenetic features of the rDNA promoter in proliferating cells and quiescent cells. In growth-arrested cells, increased levels of antisense transcripts, termed PAPAS (promoter and pre-rRNA antisense), recruit the H4K20 methyltransferase Suv4–20h2 to the rDNA promoter, thereby triggering H4K20 trimethylation and chromatin compaction. Increased levels of H4K20me3 are accompanied by decreased histone H4 acetylation, whereas H3K4 methylation is not affected.
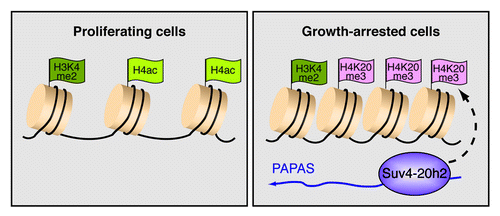
Figure 3. RNA shapes the epigenetic landscape at centromeres. The model shows the chromatin organization of minor and major satellite repeats at murine centromeres. Minor satellites in the centric region are characterized by alternating nucleosomes that harbor either the histone H3 variant CENP-A or H3K4me2. Transcripts in both orientations facilitate the association with centric proteins including CENP-C, INCENP, Survivin and aurora B kinase (AUKB). At pericentric heterochromatin, major satellite transcripts mediate the deposition of HP1α on nucleosomes marked by H3K9me3 and H4K20me3.
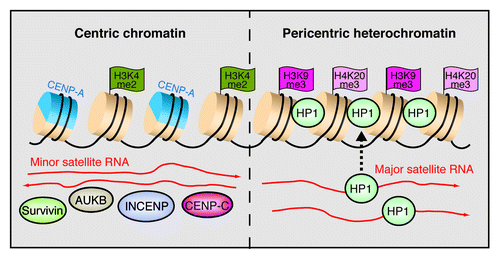
Figure 4. TERRA recruits NoRC to telomeres to establish/maintain heterochromatin at chromosome ends. TIP5 interacts with histone deacetylases (HDACs) and histone methyltransferases (HMTs), leading to deacetylation of histone H4 and trimethylation of H3K9 and H4K20.
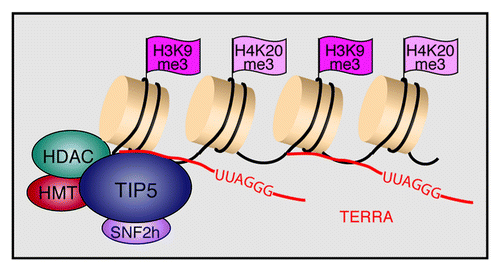
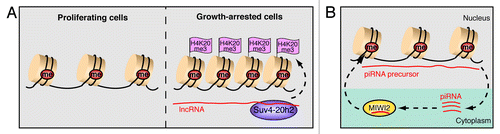
Figure 5. Non-coding RNA silences transposable elements in somatic and germ cells. (A) Somatic cells suppress transposon activity by DNA methylation and acquire H4K20 trimethylation (H3K20me3) upon growth arrest. Heterochromatin formation in quiescent cells involves upregulation of transposon-derived lncRNAs that guide Suv4–20h2 to the locus and induce H4K20me3. (B) Retrotransposons are repressed by piRNA-induced DNA methylation in murine fetal germ cells. Binding of the Piwi protein MIWI2 to mature piRNAs that originate from retrotransposon transcripts (piRNA precursors) facilitates nuclear localization and MIWI2/piRNA-dependent gene silencing.