Figures & data
Figure 1. Sequence alignment is shown for the 3XFLAG-tagged human ZFP57 aligned with three predicted human ZFP57 proteins in the UniGene database. We constructed a cDNA encoding human ZFP57 protein with a 3XFLAG tag (hZFP57–3XFLAG) at the carboxyl end. This hZFP57–3XFLAG fusion protein has an identical amino acid sequence with the predicted human ZFP57 protein NM_001109809 in the GenBank database until it reaches the junction of the 3XFLAG tag. The protein sequence for hZFP57–3XFLAG was aligned with three predicted human ZFP57 proteins (NM_001109809, BC157878 and BC171888) in the GenBank UniGene database. Shaded amino acids indicate three amino acids that are different in one of the three predicted proteins in the GenBank database. The amino acid positions for three mutations found in human patients are marked by an asterisk (*). Substitution mutations in bold letters are underlined and shown right above the mutated amino acids in the wild-type human ZFP57 protein. R248H, H277N, and H458D correspond to R228H, H257N, and H438D, respectively, that were reported in the original paper.Citation9 The amino acids from D538 to K559 for the 3XFLAG tag are indicated by bold letters at the carboxyl end of hZFP57–3XFLAG.

Figure 2. Wild-type and mutant human ZFP57 proteins were expressed in mouse ES cells lacking endogenous mouse ZFP57. (A) Schematic diagrams are shown for the wild-type human ZFP57 (hZFP57) and mouse ZFP57 (mZFP57) proteins. Same colors indicate the conserved domains with high amino acid sequence identities. Arrows with corresponding amino acid substitutions above them indicate the positions of three point mutations found in human patients. Both hZFP57 and mZFP57 contain a highly conserved KRAB box (in blue) at the N-terminus. The full-length hZFP57 is predicted to have seven zinc finger (ZF) domains. R248H and H277N are located in ZF3 and ZF4 of hZFP57 (both in red), respectively. H458D is located in the last zinc finger ZF7 of hZFP57 (in orange). By contrast, mouse ZFP57 (mZFP57) protein only has five zinc finger domains (ZF1-ZF5). ZF3 and ZF4 of hZFP57 are highly homologous to ZF2 and ZF3 of mZFP57. ZF1, ZF5, and ZF7 of hZFP57 share high sequence similarities to the corresponding ZF1, ZF4 and ZF5 of mZFP57. (B) Western blot analysis of the whole-cell lysate isolated from the Zfp57-null mouse ES clones expressing the wild-type human ZFP57 (hZFPWT) or one of three mutant human ZFP57 proteins (R248H, H277N and H458D) with a 3XFLAG tag at the C-terminus. Lanes 1–3, three independent Zfp57-null mouse ES clones expressing the wild-type human ZFP57 (hZFPWT). Lanes 4–5, two independent Zfp57-null mouse ES clones expressing the R248H mutant human ZFP57 protein. Lanes 6–7, two independent Zfp57-null mouse ES clones expressing the H277N mutant human ZFP57 protein. Lanes 8–9, two independent Zfp57-null mouse ES clones expressing the H458D mutant human ZFP57 protein. Top panel, western blot probed with mouse monoclonal anti-FLAG antibody. Bottom panel, western blot probed with mouse monoclonal antibody against β-actin.
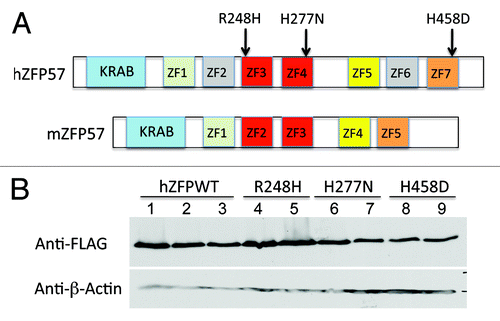
Figure 3. COBRA analysis indicates that wild-type but not mutant human ZFP57 can substitute for endogenous mouse ZFP57 in maintaining DNA methylation imprint at three imprinted regions in mouse ES cells. The mouse ES cell system we constructed before was used to drive expression of wild-type or mutant human ZFP57–3XFLAG proteins in the Zfp57-null mouse ES cells.Citation11 Exogenous human ZFP57–3XFLAG protein was constitutively expressed from an integrated transgene at the hprt locus after Cre recombinase-mediated recombination.Citation11 Simultaneously, expression of the endogenous mouse ZFP57 was turned off after excision of both floxed alleles of the mouse Zfp57 gene. Genomic DNA was isolated from these ES clones grown on gelatin-coated plates before being subjected to COBRA. Lanes 1–4, four independent Zfp57-null mouse ES clones expressing the wild-type human ZFP57–3XFLAG protein (hZFPWT). Lanes 5–8, four independent Zfp57-null mouse ES clones expressing the R248H mutant human ZFP57–3XFLAG protein. Lanes 9–12, four independent Zfp57-null mouse ES clones expressing the H277N mutant human ZFP57–3XFLAG protein. Lanes 13–16, four independent Zfp57-null mouse ES clones expressing the H458D mutant human ZFP57–3XFLAG protein. Lanes 17–18, two independent control (Ctrl) Zfp57-null mouse ES clones expressing mouse ZFP57 tagged with a Myc epitope and six histidines at the C-terminal end. Lane 19, parental (P) mouse ES cells with two floxed alleles of Zfp57. U, unmethylated bisulphite PCR product after restriction enzyme digestion. M, methylated bisulphite PCR product after restriction enzyme digestion. (A) COBRA analysis of the IG-DMR region at the Dlk1-Dio3 imprinted domain with TaqI digestion. (B) COBRA analysis of the Snrpn imprinted region with HhaI digestion. (C) COBRA analysis of the Zac1 imprinted region with TaqI digestion. (D) PCR genotyping of the endogenous mouse Zfp57 alleles in the ES clones.Citation8 Flox, the floxed allele of Zfp57 with two LoxP sites flanking two exons of Zfp57.Citation8 As expected, parental cells in Lane 19 contain two floxed alleles. One ES clone expressing R248H mutant human ZFP57 in Lane 5 also has some ES cells containing a floxed allele of mouse Zfp57. Del, the deleted allele of Zfp57 after Cre recombinase-mediated excision of the floxed allele of Zfp57.Citation8 Asterisk (*), the wild-type allele of mouse Zfp57. It appears that a small fraction of the cells were the feeder cells carried over with the ES cells when the ES clones were plated on gelatin-coated plates for genomic DNA preparation.
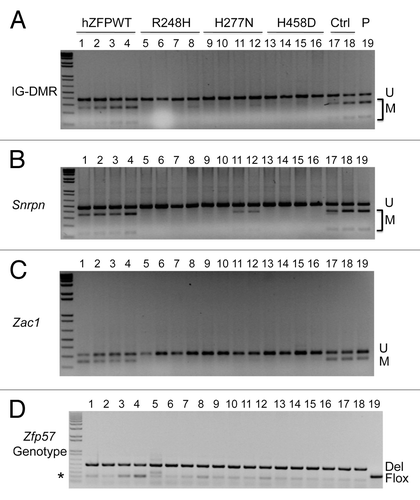
Figure 4. Wild-type and mutant human ZFP57 proteins bound to mouse KAP1 in mouse ES cells. (A) Co-immunoprecipitation (co-IP) was performed for endogenous mouse KAP1 and constitutively expressed exogenous human wild-type or one of the mutant ZFP57 proteins in mouse ES cells. ES cells were lysed in RIPA buffer for co-IP. Mouse monoclonal anti-FLAG M2 affinity gel (Sigma catalog #A2220) was used for immunoprecipitation (IP) to pull down other proteins associated with constitutively expressed exogenous wild-type or mutant human ZFP57 proteins in mouse ES cells. The immunoprecipitate was subjected to western blot (WB) analysis with affinity-purified rabbit polyclonal antibodies against mouse KAP1.Citation20 The immunoprecipitate was also probed with mouse anti-FLAG monoclonal antibody (Sigma catalog #F1804). An asterisk indicates the position of immunoglobulin (Ig). An arrow marks the position of the wild-type or mutant human ZFP57 proteins with the 3XFLAG tag. The input samples were probed with these two antibodies as well as the mouse monoclonal anti-β-actin antibody (Sigma catalog #A1978). Lane 1, a Zfp57-null ES clone 2B6.Citation8,Citation11 Lane 2, an ES clone expressing wild-type human ZFP57 (hZFPWT). Lane 3, an ES clone expressing R248H mutant human ZFP57–3XFLAG protein. Lane 4, an ES clone expressing H277N mutant human ZFP57–3XFLAG protein. Lane 5, an ES clone expressing H458D mutant human ZFP57–3XFLAG protein. (B) Wild-type and mutant human ZFP57 proteins exhibited similar binding affinities for KAP1 in mouse ES cells. The intensities of each band in the western blot were measured by Image J. Relative binding affinities were calculated based on the ratios of the band intensity of anti-KAP1 over the band intensity of anti-FLAG for each ES sample in the co-IP western blot in A, with the binding affinity of the wild-type human ZFP57 for mouse KAP1 set at 1. We reason that mouse KAP1 is much more abundant than human ZFP57 in mouse ES cells and KAP1 was not a limiting factor in the co-IP assay. Thus, we did not take the concentration of KAP1 in various ES clones into account in estimating the relative binding affinities between mouse KAP1 and human ZFP57 proteins.
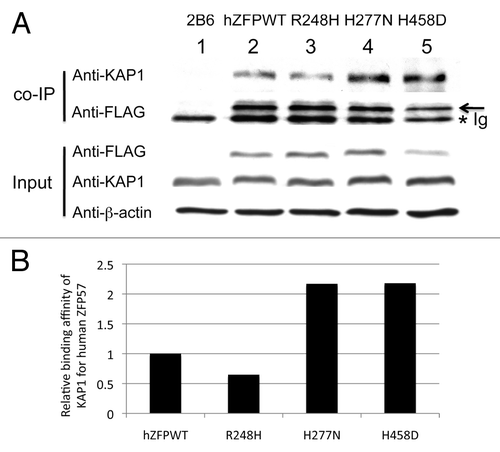
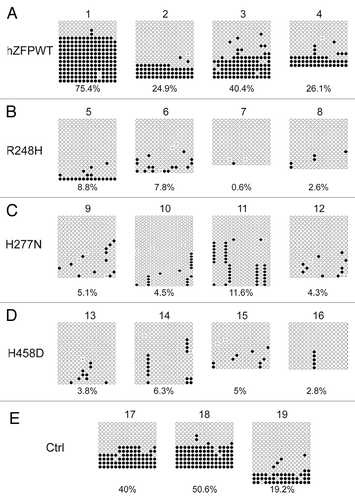
Figure 5. Bisulphite sequencing result confirms that wild-type human ZFP57 can substitute for endogenous mouse ZFP57 in maintaining DNA methylation imprint at the IG-DMR of the Dlk1-Dio3 imprinted region in mouse ES cells. Genomic DNA samples from the ES clones plated on the gelatin-coated plates were subjected to bisulphite mutagenesis, PCR amplification and bacterial colony sequencing. Filled circle, a methylated CpG. Unfilled circle, an unmethylated CpG. Cross (X), a CpG site that its methylation status cannot be determined. Each row stands for a template DNA molecule directly sequenced from a single bacterial colony containing the bisulphite PCR product. Numbers 1 to 19 indicate the same ES clones that were subjected to COBRA analysis in . Percentage of methylated CpG sites in all sequenced bacterial colonies for each ES clone is listed directly below the diagram illustrating methylated and unmethylated CpG sites. (A) Four independent Zfp57-null mouse ES clones expressing the wild-type human ZFP57–3XFLAG protein (hZFPWT). (B) Four independent Zfp57-null mouse ES clones expressing the R248H mutant human ZFP57–3XFLAG protein. (C) Four independent Zfp57-null mouse ES clones expressing the H277N mutant human ZFP57–3XFLAG protein. (D) Four independent Zfp57-null mouse ES clones expressing the H458D mutant human ZFP57–3XFLAG protein. (E) control (Ctrl) ES clones. 17 and 18, two independent Zfp57-null mouse ES clones expressing mouse ZFP57 tagged with a Myc epitope and six histidines at the C-terminal end. 19, parental (P) mouse ES cells with two floxed alleles of Zfp57.
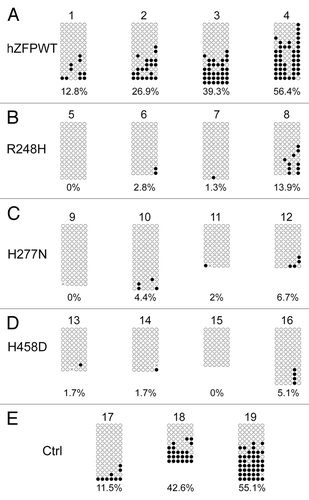
Figure 6. Bisulphite sequencing result confirms that wild-type human ZFP57 can substitute for endogenous mouse ZFP57 in maintaining DNA methylation imprint at the Snrpn DMR region in mouse ES cells. Genomic DNA samples from the ES clones plated on the gelatin-coated plates were subjected to bisulphite mutagenesis, PCR amplification and bacterial colony sequencing. Filled circle, a methylated CpG. Unfilled circle, an unmethylated CpG. Cross (X), a CpG site with unknown methylation status. Each row stands for a template DNA molecule directly sequenced from a single bacterial colony containing the bisulphite PCR product. Numbers 1 to 19 indicate the same ES clones that were subjected to COBRA analysis in . Percentage of methylated CpG sites in all sequenced bacterial colonies for each ES clone is listed directly below the diagram illustrating methylated and unmethylated CpG sites. (A) Four independent Zfp57-null mouse ES clones expressing the wild-type human ZFP57–3XFLAG protein (hZFPWT). (B) Four independent Zfp57-null mouse ES clones expressing the R248H mutant human ZFP57–3XFLAG protein. (C) Four independent Zfp57-null mouse ES clones expressing the H277N mutant human ZFP57–3XFLAG protein. (D) Four independent Zfp57-null mouse ES clones expressing the H458D mutant human ZFP57–3XFLAG protein. (E) Control (Ctrl) ES clones. Numbers 17 and 18, two independent Zfp57-null mouse ES clones expressing mouse ZFP57 tagged with a Myc epitope and six histidines at the C-terminal end. 19, parental (P) mouse ES cells with two floxed alleles of Zfp57.
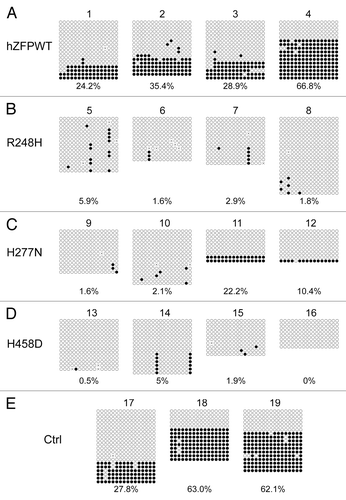
Figure 7. Bisulphite sequencing result confirms that wild-type human ZFP57 can substitute for endogenous mouse ZFP57 in maintaining DNA methylation imprint at the Zac1 DMR region in mouse ES cells. Genomic DNA samples from the ES clones plated on the gelatin-coated plates were subjected to bisulphite mutagenesis, PCR amplification and bacterial colony sequencing. Filled circle, a methylated CpG. Unfilled circle, an unmethylated CpG. Cross (X), a CpG site with unknown methylation status. Each row stands for a template DNA molecule directly sequenced from a single bacterial colony containing the bisulphite PCR product. Numbers 1 to 19 indicate the same ES clones that were subjected to COBRA analysis in . Percentage of methylated CpG sites in all sequenced bacterial colonies for each ES clone is listed directly below the diagram illustrating methylated and unmethylated CpG sites. (A) Four independent Zfp57-null mouse ES clones expressing the wild-type human ZFP57–3XFLAG protein (hZFPWT). (B) Four independent Zfp57-null mouse ES clones expressing the R248H mutant human ZFP57–3XFLAG protein. (C) Four independent Zfp57-null mouse ES clones expressing the H277N mutant human ZFP57–3XFLAG protein. (D) Four independent Zfp57-null mouse ES clones expressing the H458D mutant human ZFP57–3XFLAG protein. (E) Control (Ctrl) ES clones. Numbers 17 and 18, two independent Zfp57-null mouse ES clones expressing mouse ZFP57 tagged with a Myc epitope and six histidines at the C-terminal end. Number 19, parental (P) mouse ES cells with two floxed alleles of Zfp57.