Figures & data
Table 1. Classification of lncRNAs.
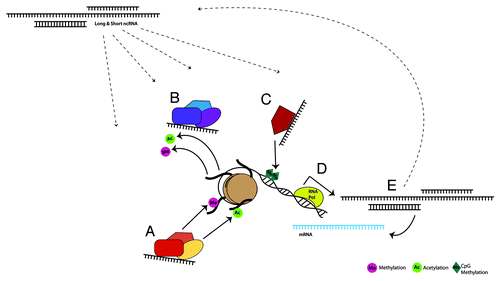
Figure 1. Environmentally sensitive, dynamic regulation of gene expression by long and short ncRNAs occurs through direct and indirect interaction with classical epigenetic mechanisms, forming a larger epigenetic network. ncRNAs are known to bind and recruit histone modifying complexes to either add (A) or remove (B) methyl and acetyl groups. These transcripts also modulate DNA methyltransferases (C), thereby facilitating or suppressing DNA methylation. Loci that are targeted by each of these mechanisms can encode protein coding and/or noncoding RNA transcripts (D). ncRNAs in turn can affect gene expression by interacting with mRNA, or through feedback into the previously discussed pathways (E).