Figures & data
Figure 1. Analysis of MIR941 and MIR1247 expression in gastric cancer cell lines. (A) Quantitative RT-PCR analysis of expression pattern of pri-MIR941 and pri-1247 in gastric cancer cell lines (KATO III, NCI-N87, AGS, and AZ521) and normal stomach tissues (NG). (B) Expression analysis of miRNAs in gastric cancer cells with 5-aza-dC treatment. RT-PCR was performed to assess pri-miRNAs levels in gastric cancer cell lines (KATO III, NCI-N87, AGS, and AZ521) before and after treatment with 5 μM 5-aza-dC for 72hrs. Dark gray bar indicate 5-aza-dC treatments. (C) Expression of mature MIR941 and MIR1247 in gastric cancer cells with 5-aza-dC treatment. RT-PCR was performed to assess mature miRNAs levels after treatment with 5 μM 5-aza-dC for 72 h. Dark gray bars indicate 5-aza-dC treatments. Relative pri-mRNAs levels are shown as mean ± standard deviation of three independent experiments. *Indicates significant increase in pri-miRNA expression after 5-aza-dC treatment (*P < 0.05).
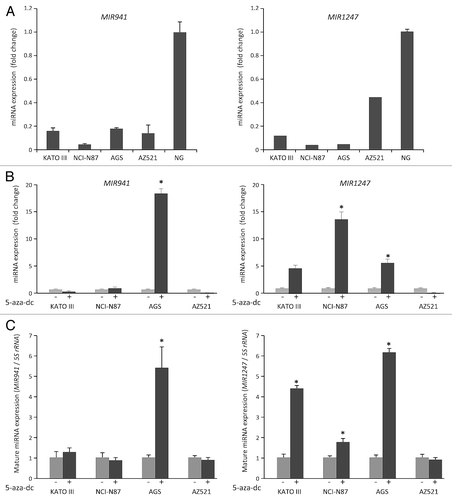
Figure 2. Methylation analysis of MIR941 and MIR1247 in gastric cancer cell lines. (A) Schematic representation of MIR941 and MIR1247 CpG island (dotted box). Both miRNAs are embedded into the CpG island (gray box). The regions analyzed using methylation specific PCR (MSP), bisulfite sequencing are indicated by black bars below the CpG island. (B) MSP analysis of MIR941 and MIR1247 in four gastric cancer cell lines. In vitro methylated DNA (IVD) and DKO cells were used as positive and negative controls, respectively. U and M indicates unmethylation and methylation signals, respectively. (C) Representative bisulfite sequencing analysis was performed for MIR941 and MIR1247 in gastric cancer cell lines (AGS, NCI-N78, and KATO III) as well as cells treated with 5-aza-dC. Each box represents a CpG dinucleotide. Black boxes represent methylated cytosines while white boxes represent unmethylated cytosines.
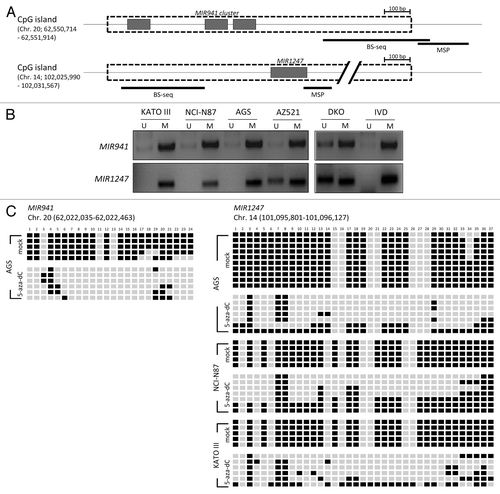
Figure 3. Correlation between expression and methylation of MIR941 and MIR1247 in gastric cancer patient samples. (A) Expression level of pri-MIR941 and pri-1247 in primary gastric tumor tissues compared with normal gastric mucosae from healthy individuals by qRT-PCR. * indicates significant decrease in miRNA expression (*P < 0.05). (B) Representative bisulfite sequencing analysis was performed for MIR941 and MIR1247 in three normal gastric tissues and five gastric adenocarcinomas. Each box represents a CpG dinucleotide. Black boxes represent methylated cytosines while white boxes represent unmethylated cytosines.
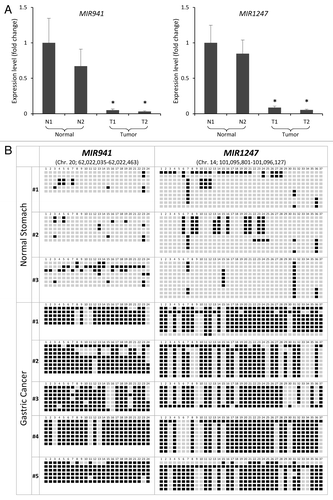
Figure 4. Functional analyses of MIR941 and MIR1247 in AGS and KATO III cells. (A) Growth curves and (B) MTT assays of AGS mock treated cells and AGS cells transfected with non-targeting negative control miRNA (All Star neg.), MIR941, or MIR1247 mimics. (C) Wound healing assay for AGS cells transfected with mock, non-targeting negative controls miRNA (All Star neg.), MIR941, or MIR1247 mimics. Photographs were taken at 16 h later after wounding. (D) The bar graph represents the quantity of migrated cells using transwell migration assay with AGS cells transfected with mock, non-targeting negative controls miRNA (All Star neg.), MIR941, or MIR1247 mimics. Quantity of migrated cells represents the mean of 3 random microscopic fields per membrane and error bars represent the SDs. (E) Transwell invasion assay using AGS cells transfected with mock, non-targeting negative controls miRNA (All Star neg.), MIR941, or MIR1247 mimics. Representative field of invasive cells on the membrane are shown. The Bar graph represents the mean of 3 random microscopic fields per membrane and error bars represent the SDs.
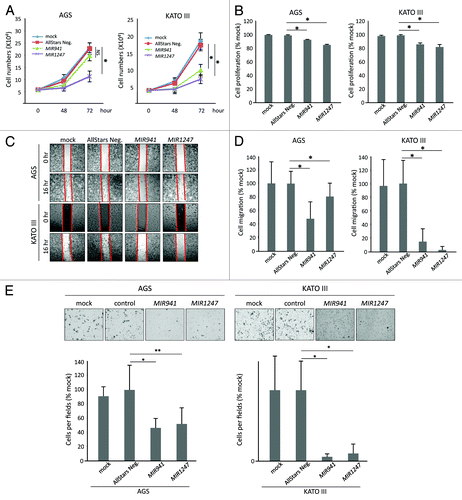
Table1. Target gene list of MIR941 and MIR1247
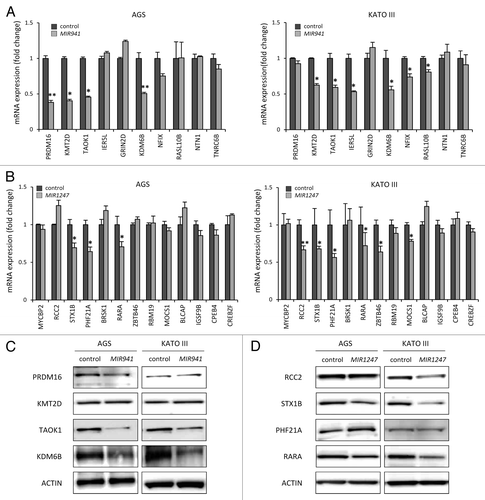
Figure 5.Candidate target genes are downregulated by MIR941 and MIR1247 mimics. (A and B) Real-time RT-PCR was performed in AGS and KATO III cells transfected with non-targeting negative control miRNA (All Star neg.), MIR941 and MIR1247 mimics with target genes listed in . *Indicates statistically significance decrease of gene expression level compared with control (P < 0.05). (C and D) Western blot analysis of target genes in AGS and KATO III cells transfected with MIR941 and MIR1247, or negative control.