Figures & data
Figure 1. LINE-1 DNA methylation in T-lymphocytes and B-lymphocytes from patients with rheumatoid arthritis. Methylation at three adjacent CpG sites within LINE-1 repetitive sequences was quantified in purified T- and B-lymphocytes from 12 RA patients by sodium bisulfite pyrosequencing. Unfilled circles and crosses represent T-lymphocytes and B-lymphocytes, respectively. The mean levels of methylation for each of the CpG sites are shown by the short black horizontal bars.* P ≤ 0.01 (Wilcoxon Signed-Rank Test).
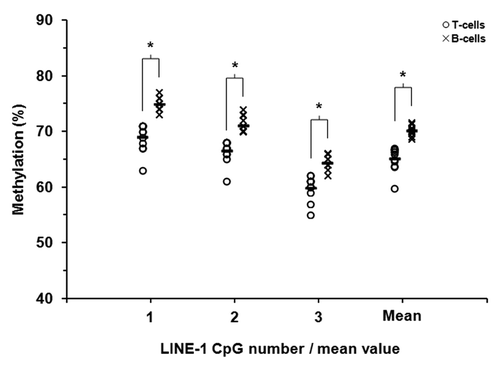
Figure 2. DNA methylation heatmap for the 679 CpG methylation signature in RA derived T- and B-lymphocytes. CpGs presented in the heatmap were described in our previous work (22) and represent those sites which define a unique methylation signature distinguishing healthy T-lymphocytes from B-lymphocytes. Each row represents an individual CpG, ordered according to the clustering output in healthy individuals (22), and each column a different sample (listed beneath the heatmap). Color gradation from yellow to blue represents low to high DNA methylation respectively, with β-values ranging from 0 (no methylation; yellow) to 1 (complete methylation; blue). Intrinsic methylation differences previously observed between the cell types in healthy individuals are accurately preserved in RA patients.
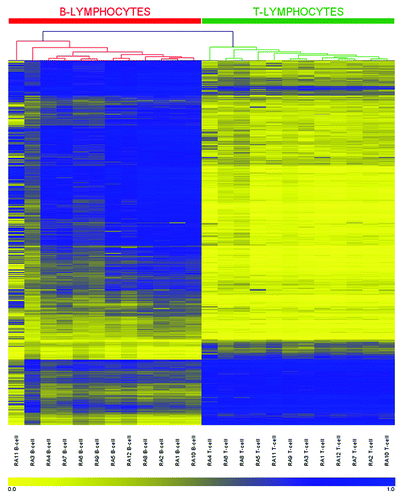
Figure 3. Filtering criteria for identification of CpGs differentially methylated between RA patients and healthy individuals. For T-lymphocytes and B-lymphocytes separately, the starting number of CpGs (477,312 and 479,639 for T- and B-lymphocytes respectively) was derived through the removal of unreliable sites (CpGs with detection p-values > 0.05) and those with missing β-values, as described in the Methods. Numbers indicate the number of CpGs remaining at each step.† Refers to CpGs for which all RA samples showing a change from the healthy mean ± two-times the SD in the preceding step were either hypermethylated or hypomethylated in RA.
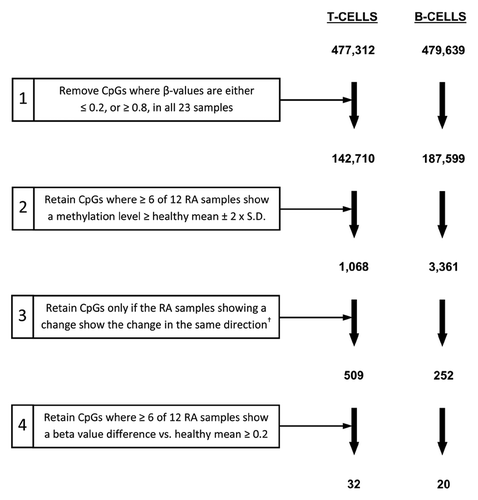
Table 1. Summary characteristics for the differentially methylated CpGs identified in RA derived T-lymphocytes and B-lymphocytes.*
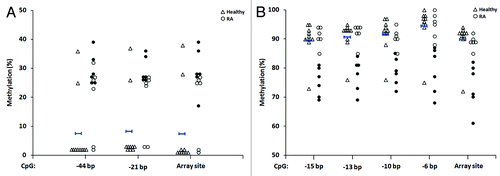
Figure 4. Validation by bisulfite pyrosequencing of differentially methylated CpG candidates in RA derived T- and B-lymphocytes. For each cell type, four candidate CpGs that were differentially methylated between RA patients and healthy individuals were selected. This included two sites each that were hypermethylated (ARSB and DUSP22) and hypomethylated (GALNT9 and MGMT) in RA derived T-lymphocytes (A), and included four hypomethylated sites in B-lymphocytes (B). In each plot, unfilled triangles indicate healthy individuals and where the mean methylation and two-times the SD of the mean are indicated for the array data by short blue horizontal bars with vertical stops and with arrowheads, respectively. RA patients are indicated by circles, where filled circles in the array data indicate the individual samples that were identified as differentially methylated relative to healthy individuals (as defined by the criteria in ). These same samples are also indicated by filled circles in the pyrosequencing data. Abbreviations: Pyro., bisulfite pyrosequencing.
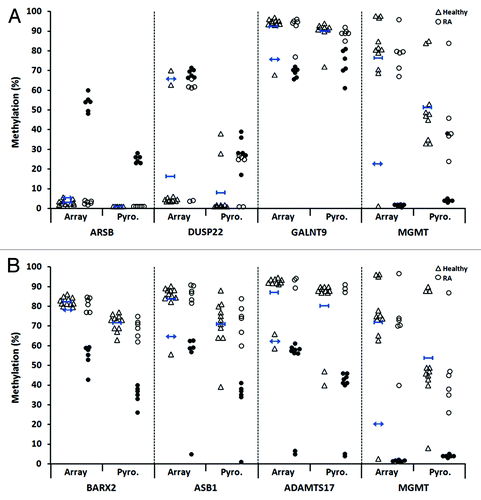
Figure 5. Identification of additional differentially methylated CpGs for two array-identified candidates in RA derived T-lymphocytes. Shown are additional CpGs adjacent to the array-identified candidate site for DUSP22 (two additional sites) (A) and GALNT9 (four additional sites) (B) that were also found to be differentially methylated in RA patients relative to healthy individuals, as determined by pyrosequencing. Negative base pair numbers on the x-axis indicate bases upstream of the array CpG site. In each plot, unfilled triangles indicate healthy individuals and where the mean methylation is indicated by short blue horizontal bars with vertical stops. RA patients are indicated by circles, where filled circles for the array site indicate the individual samples that were identified as differentially methylated relative to healthy individuals (as defined by the criteria in ). These same samples are also indicated by filled circles in each of the adjacent CpGs presented. Abbreviations: bp, base pairs.