Figures & data
Figure 1. AS-ChIP reveals high resolution maps of allelic imbalance for 5 histone modifications (A) Twenty cell lines from 3 cohorts comprising 2 distinct cell-types, 2 ancestral populations and 3 parent-child trios underwent AS-ChIP analysis. Fibroblast (FB) cell-lines came from 2 trios of Caucasian ancestry. LCL cell-lines came from 2 ‘1000 Genomes’ populations, CEU (Utah residents of European ancestry) and YRI (Individuals of Yoruban ancestry in Ibadan, Nigeria). From each of these two populations we selected 1 trio and 4 unrelated children (for which all parents’ phased genotypes were available for phasing). Ovals indicate females and rectangles indicate males. (B) Chromatin immunoprecipitation for each of the 5 histone modifications: H3K4me1 (enhancer), H3K4me3 (promoter), H3K27me3 (inactive chromatin), H3K27ac (active chromatin), H3K36me3 (transcriptional elongation). ChIP samples were run along with cDNA and gDNA from each cell-line, on high density Illumina genotyping arrays, covering ~4.4 million common SNPs. By assessing the signal ratio coming from the X and Y channel for each SNP we were able to calculate allelic imbalance values for heterozygous SNPs. (C) An example of AS-ChIP AI data for the paternally-expressed imprinted gene PLAGL1 in the fibroblast cell-line GM02456, shown on the UCSC genome browser track. Red upwards bars correspond to AI in favor of the paternal allele at the given SNP, with blue bars indicating AI in favor of the maternal allele.
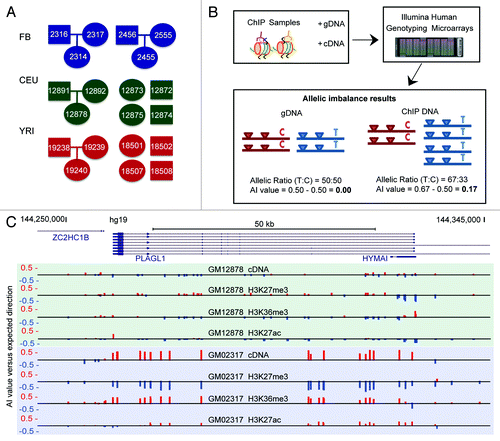
Table 1. Number of heterozygous SNPs with minimum AI of 0.05 by assay
Figure 2. Global characterization of allelic chromatin modifications around transcripts with differential allelic expression Allelic imbalance plotted for the 5 histone modifications across 3513 transcripts demonstrating allelic expression: (A) H3K4me1, (B) H3K4me3, (C) H3K27ac, (D) H3K36me3, (E) H3K27me3. Colored lines indicate average AI calculated using AI data for 25 bins of 10 kb, 2 kb upstream of TSS, 1st exon, 1st intron (1st 2 kb max), rest of transcript, last exon, 2 kb downstream of TES, and 25 bins of 10 kb further upstream of TSS. Grey plots show the –log (P value) for each bin, calculated via T-test, using as a baseline AI value the mean AI of the 2bins at the 250 kb upstream and downstream of TSS and TES respectively. Gray dotted lines indicate where P = 0.05 (corrected for multiple testing).
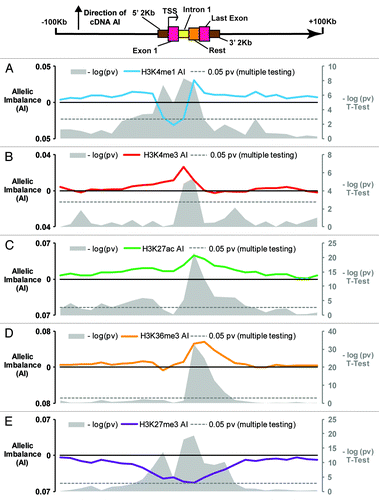
Table 2. Significant allelic bias validation using Sanger sequencing
Figure 3. H3K4me1 and H3K4me3 allelic chromatin domains pairwise overlap demonstrates cell-type specificity, population-specificity and heritability (A–B) Heat maps of enrichment of pair-wise overlap between individuals for (A) H3K4me1 and (B) H3K4me3 allelic-chromatin domains, vs. shuffled control regions of equal size. Along the top and side axes are each of the AS-ChIP individuals, labeled by population and whether they are in a trio by their relationship: daughter (D), mother (M), father (F), or unrelated (U). (C-D) Bar charts for H3K4me1 (C) and H3K4me3 (D), showing mean enrichment for pair-wise overlap is significantly higher within cell-types among unrelated individuals from same population (purple) vs. across cell-types. Also pair-wise overlap is significantly higher among unrelated individuals within the same population (purple), vs. across populations, same cell-type, unrelated individuals (green). Finally pair-wise overlap is significantly higher among related individuals from same cell-type and population (purple), vs. unrelated individuals from same cell-type and population (green). * = P < 0.01, ** = P < 0.001, *** = P < 10E-5.
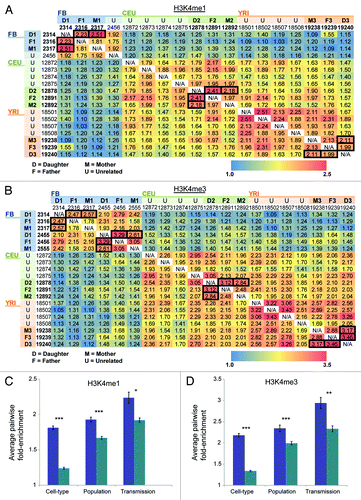
Figure 4. dsQTL–positive DNase peaks preferentially overlap allelic chromatin domains Enrichment of DHS peaks in H3K4me1 and H3K4me3 allelic-chromatin domains from each of the 3 populations (CEU, FB and YRI). Dark blue indicates dsQTL positive DHS peak overlap, and light blue indicates dsQTL negative DHS peaks overlap. n.s = P > 0.05, ** = P < 0.001, *** = P < 10E-5.
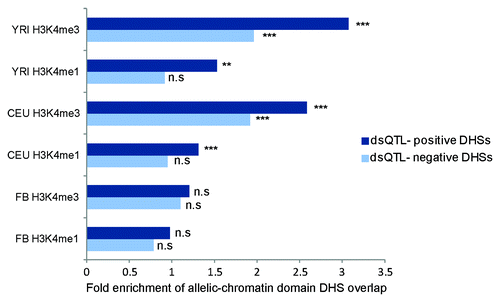
Figure 5. Enrichment of disease-associated variants in allelic chromatin domains. Enrichment of GWAS lead SNPs from GWAS catalog in allelic-chromatin domains, vs. control regions of equal size. Green, red and blue bars indicate YRI, CEU and FB and populations respectively with darker bars being H3K4me3 domains and pale bars being H3K4me1 domains. n.s = P > 0.05, * = P < 0.01, ** = P < 0.001, *** = P < 10E-5.
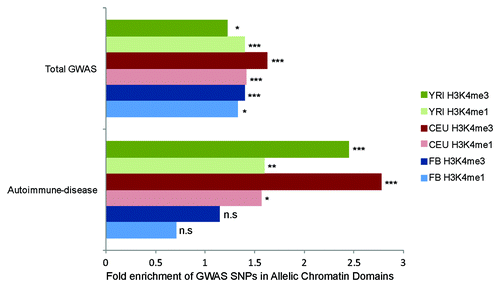
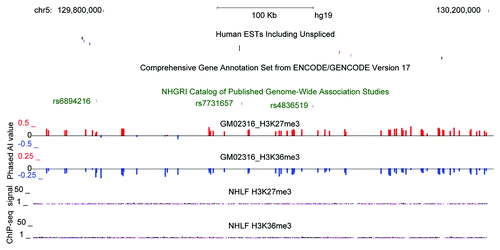
Figure 6. Allelic-chromatin suggests allelic transcription in an unannotated region overlapping GWAS SNPs Example of H3K27me3 and H3K36me3 allelic-chromatin suggestive of transcription in intergenic region of genome on chromosome 5 in UCSC genome browser for GM02316. Red and blue bars correspond to phased AI in opposite directions. Tracks above show all human ESTs in the region, a lack of any annotation in GENCODE V17 Comprehensive annotation and 3 GWAS SNPs, in the GWAS catalog. Below are ChIP-seq tracks from ENCODE showing low signal for H3K27me3 and H3K36me3 in NHLF (Normal Human Lung Fibroblast).