Figures & data
Figure 1 Cellular proliferation demonstrated by BrdU incorporation into HCT116 p53+/+ (A) and p53-/- (B) cells grown under control, no oxygen and no glucose conditions. Arrows indicate cells which are proliferating (BrdU positive), while arrow heads indicate BrdU negative cells. Scale bar represents 100 µm. (C) We examined apoptosis under ischemic conditions via western blotting for caspase-3 cleavage. Top: blot shows no detectable cleaved caspase-3 in any of the samples. Bottom: the same blot after prolonged exposure reveals low amounts of cleaved (activated) caspase-3 in p53-/- cells exposed to no glucose conditions.
Figure 2 Quantitative real-time reverse transcription PCR. To represent mRNA, cDNA levels of DNMT1 (A), DNMT3a (B) and DNMT3b (C) from p53+/+ and p53-/- HCT116 cells, after 24 h in control (C), low oxygen (LO) and low glucose (LG). mRNA levels were normalized to α-actin expression. DNMT expression of each cell line was normalized to the control condition. Averages of 5 independent experiments with standard error are shown. *Indicates p ≤ 0.05 relative to cell type respective control. **Indicates p ≤ 0.05 relative to low oxygen.
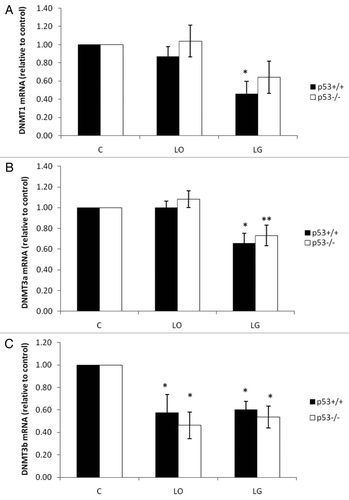
Figure 3 Protein expression profiles of DNMT1 and DNMT3a. Protein levels of DNMT1 (A and B) and DNMT3a (C and D) in p53+/+ and p53-/- HCT116 cells cells after 24 h in control (C), low oxygen (LO) and low glucose (LG). DNMT1 (200 kDa) and DNMT3a (130 kDa) protein levels were normalized to α-tubulin (50 kDa) to account for any protein loading discrepancies. Averages of three independent experiments with standard error are shown. *indicates p ≤ 0.05 relative to cell type respective control.
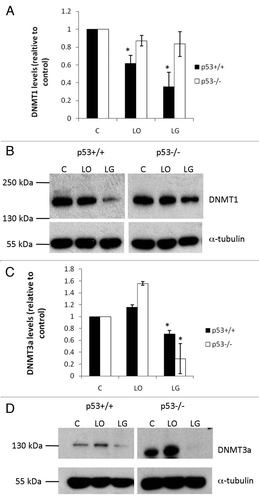
Figure 4 Total DNMT activity. Activity levels of total DNMTs were measured with EpiQuik™ DNA Methyltransferase Activity/Inhibition Assay Kit. Nuclear protein was isolated from p53+/+ and p53-/- HCT116 cells after 24 h in control (C), low oxygen (LO) and low glucose (LG). The assay measures the amount of DNA methylation, which is proportional to DNMT activity. The average of four independent experiments with standard error is shown. *indicates p ≤ 0.05 relative to cell type respective control.
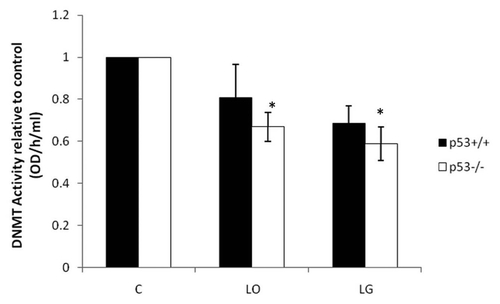
Figure 5 Detection of DNMT1 and hypoxic regions in HCT116 xenografts by immunofluorescence. Blue channel is nuclear staining by DAPI (A), red channel is DNMT1 (B), green is CA IX/hypoxia marker (C), and overlay of DNMT1 and CA IX (D). Arrows indicate cells which are hypoxic and DNMT1 negative. Asterisks indicate cells which are both hypoxic and expressing DNMT1. Scale bar represents 50 µm.
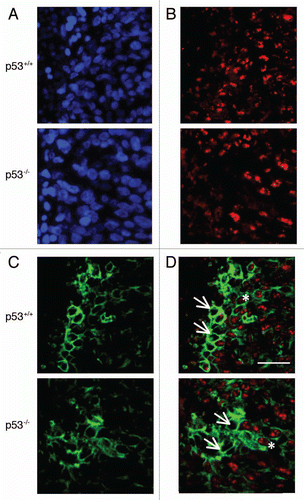
Figure 6 Western blot showing recovery of DNMT1 protein levels in HCT116 cells after 24 h in no oxygen or no glucose. Protein was collected after cells were restored to normal conditions for 0, 4, 8, 12 and 24 h.
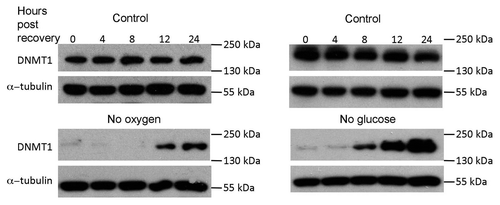
Figure 7 Map of proximal p16INK4a promoter region chosen for analysis; each circle represents a CpG (A). Detection of DNA methylation changes using bisulfite sequencing in human p16INK4a promoter region following exposure to ischemia in vitro for 48 h (B). Open and closed circles represent unmethylated and methylated CpGs, respectively. A dash is an unknown methylation status. In p53+/+ cells, there were significant changes in p16INK4a methylation after both no oxygen and no glucose treatments (p ≤ 0.05). In p53-/- cells, there was a significant decrease in methylation following no glucose treatment (p ≤ 0.05).
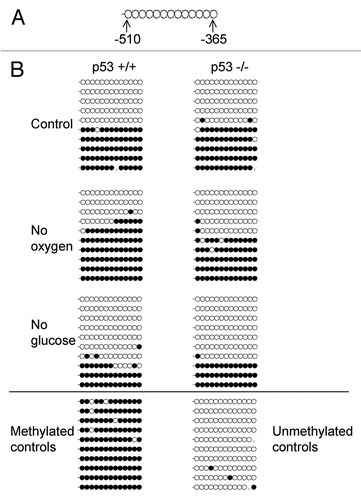
Table 1 Primers and sequences used in quantitative real-time PCR and bisulfite sequencing