Figures & data
Figure 1 The Avy metastable epiallele. (A) The viable yellow agouti (Avy) allele contains a contra-oriented IAP insertion within pseudoexon 1A (PS1A) of the Agouti gene. A cryptic promoter (short arrow labeled Avy) drives constitutive ectopic Agouti expression. Transcription of the Agouti gene normally initiates from a developmentally regulated hair-cycle specific promoter in exon 2 (short arrow labeled A, a). (B) Genetically identical Avy/a offspring representing the yellow and pseudoagouti coat color phenotypes are shown. * indicates 5′LTR of the Avy IAP region and **represents non-IAP genomic PS1A region.
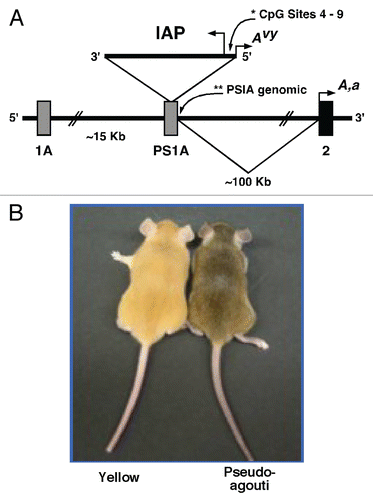
Figure 2 Chromatin precipitation for acetylated histones H3 and H4 in the 5′ LTR of the IAP and in PS1A. Binding activity was calculated as percent of pre-immunoprecipitated input DNA as represented by 2−ΔC(t) X100. (A) DNA precipitated by H3 di-acetylation antibody is enriched in yellow versus pseudoagouti Avy/a mice (p = 0.09; n = 6 per group) within the 5′ LTR of the IAP. (B) DNA precipitated by H4 di-acetylation antibody is enriched in yellow versus pseudoagouti Avy/a mice (p = 0.08; n = 3 per group) within the 5′ LTR of the IAP. (C) DNA precipitated by H3 di-acetylation antibody does not vary by coat color within the PS1A genomic region (p = 0.4; n = 6 per group). (D) DNA precipitated by H4 di-acetylation antibody does not differ by coat color within the genomic PS1A region (p = 0.4; n = 3 per group). * indicates significance at the 0.09 level.
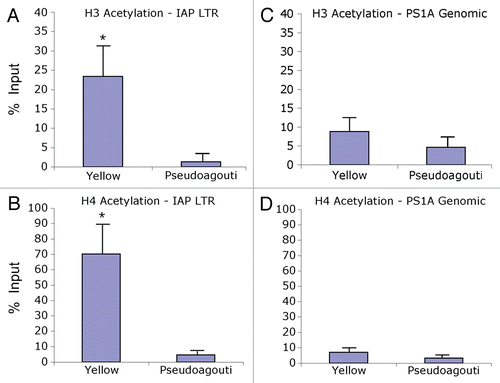
Figure 3 Chromatin precipitation for methylated histones in the 5′ LTR of the IAP and in PS1A. Binding activity was calculated as percent of pre-immunoprecipitated input DNA as represented by 2−ΔC(t) X100. (A) DNA precipitated by H3K4 tri-methylation antibody is not enriched in yellow versus pseudoagouti Avy/a mice (p = 0.7; n = 6 per group) within the 5′ LTR of the IAP. (B) DNA precipitated by H4K20 tri-methylation antibody is enriched in pseudoagouti compared to yellow Avy/a mice (p = 0.01; n = 6 per group) within the 5′ LTR of the IAP. (C) DNA precipitated by H3K4 tri-methylation antibody does not vary by coat color within the PS1A genomic region (p = 0.9; n = 5 per group). (D) DNA precipitated by H4K20 tri-methylation antibody does not differ by coat color within the genomic PS1A region (p = 0.8; n = 5 per group). ** indicates significance at the 0.01 level.
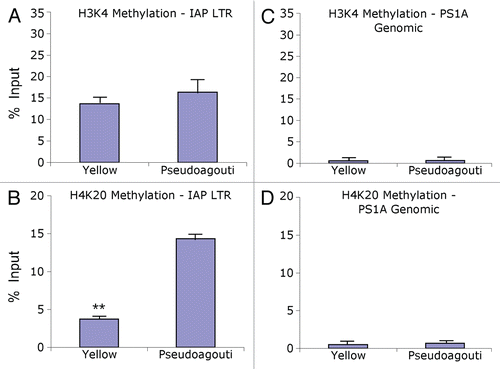
Table 1 DNA methylation and Agouti (a) expression levels by histone and coat color status
Table 2 PCR and pyrosequencing primers