Figures & data
Figure 1 SETDB1 binds histone H3 in vitro and in vivo. (A) A graphical depiction of SETDB1 functional domains. Of note, the SET domain (orange) is interrupted by a so-called bifurcation region (green) of unknown function. (B) GST-pulldown experiment. Full-length recombinant GST-SETDB1 was incubated with calf thymus histones. GST alone and GST-ING2PHD were used as negative and positive controls, respectively. Following SDS-PA GE separation and transfer, the membranes were probed with α-H3, α-H4 or α-H2A antibodies. Input lane is 5%. (C) α-H3 ppChIP. Chromatin was immunoprecipitated with α-H3 and analyzed by immunoblotting using α-H3, α-H4 and α-SETDB1. Input level for α-H3 and α-H4 is 1 and 10% for α-SETDB1. (D) Mapping of SETDB1 minimal region necessary and sufficient for H3-binding. Biotinylated histone H3 peptides were incubated with recombinant SETDB1 and streptavidin-sepharose beads used to pulldown the peptides and interacting proteins. Following SDS-PAGE separation and transfer, the membranes were probed with an HRP-conjugated α-GST antibody. (E) Representation of the truncation mutants used in (D) and a summary table of their respective affinity for H3 binding.
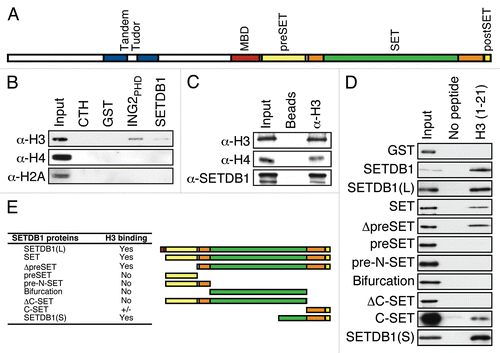
Figure 2 H3K4me3 euchromatic mark impedes SETDB1 binding in vitro. (A) Pulldowns using biotinylated H3 peptides with the indicated modifications as in . (B) A similar experiment as in (A), but ING4PHD was used in parallel as a control for H3K4me3 binding. (C) G9A and SUV39H1 were used in a biotin-pulldown experiment.
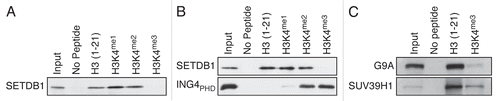
Figure 3 H3K4me3 euchromatic mark hinders SETDB1 in vitro H3 methylation. (A) The indicated histone peptides were incubated with SETDB1 and 3H-SA M, then pulled-down with streptavidin beads in 300 mM NaCl buffer and washed three times. The beads were then transferred to liquid scintillation vials for quantification of incorporated methyl groups. (B) SETDB1 and G9A were tested for KMT activity on the indicated biotin-peptides using a Flashplate. The values are in (%) with the activity on H3 (1–21) set at 100%. The error bars represent the S.E.M. of quadruplicates from a representative experiment. (C) Methyltransferase reactions using either SETDB1, G9A or SUV39H1 were conducted on full-length recombinant histone H3 with the indicated MLA modifications. The samples were analyzed by autoradiography (3H-SA M) and western blotting using α-H3K9me3. Either α-H3 or α-GST antibodies were used as controls for substrate and enzyme input in each reaction. (D) KMT activity of wild-type G9A900 and mutants C1115A, D1074A, D1074A/D1088A, D1074Y and D1088Y.
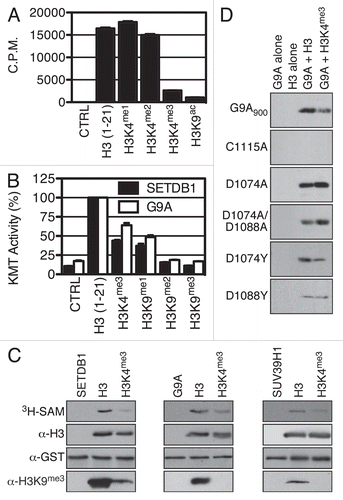
Figure 4 H3K4me2/3 and H3K9me2/3 interplay in vivo. (A) HP1γ was immuno-precipitated from cross-linked chromatin and bound histone analyzed by MS. (B) Table of the raw values obtained for (A). (C) HeLa cells were transfected with either siCTRL, siWDR82 or siRBBP5, the RNA isolated and the level of WDR82 and RBBP5 transcripts analyzed by qPCR. (D) Immunoblot analysis of H3K4me3 in cells from (C). (E) Heatmap of quantitative mass spectrometric analysis of total histones from cells in (C). (F) Raw values used to generate (E) heatmap.
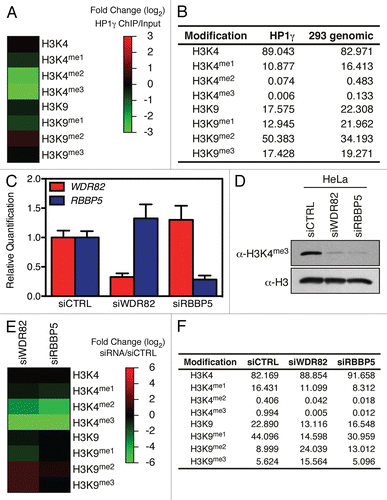
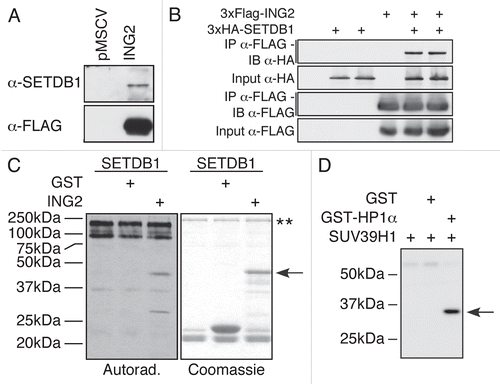
Figure 5 KMTs interact with their substrate. SETDB1 interacts with ING2. (A) FLAG-purified ING2 complex from a stably expressing cell line was probed for the presence of SETDB1. The SETDB1 antibody detected the presence of the endogenous enzyme only in the ING2 complex, but not in the control lane. (B) HEK-293T cells were transfected with an empty vector or 2 independent clones of HA-SETDB1 in the absence or presence of FLAG-ING2. The cells were lysed and proteins immunoprecipitated using FLAG-M2 agarose. The samples were separated by SDS-PAGE, transferred to PVDF and analysed with either α-HA or α-FLAG M2-HRP antibodies. (C) ING2 is methylated in vitro by SETDB1. Recombinant SETDB1 purified from insect cells was incubated alone or with either GST or GST-ING2 in the presence of 3H-SA M. Autoradiography of the experiment (left) and Coomassie staining of the gel (right). The arrow indicates GST-ING2. The two stars (**) indicate SETDB1 and its auto-methylated forms. (D) HP1α is methylated in vitro by SUV39H1. Autoradiography of an enzymatic reaction using bacterially expressed recombinant SUV39H1 and HP1α. In the first lane, only the KMT is present. In the second lane, GST was used as a negative control for SUV39H1 methylation. In the last lane, both SUV39H1 and HP1α were incubated. The arrow indicates HP1α.