Figures & data
Table 1. Biological and molecular features of CLL patients included in the study
Table 2. Global DNA methylation levels in healthy subjects compared with CLL patients
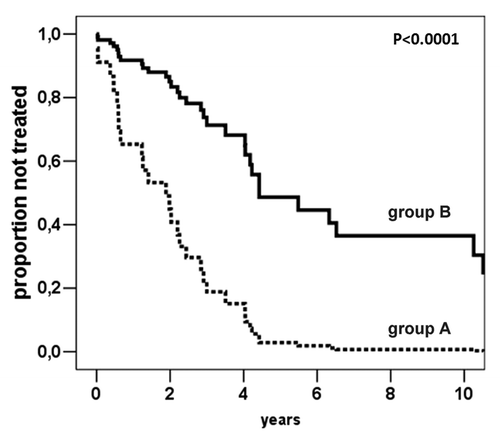
Figure 1. DNA methylation levels in relation to different cytogenetic groups. Box plot representation of DNA methylation levels. The p values corresponding to each methylation marker are shown.
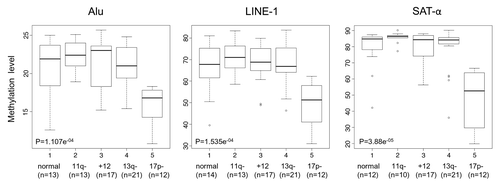
Table 3. DNA methylation levels related to different biological markers in CLL patients
Figure 2. Boxplot representation of absolute RNA expression levels of the three DNMTs(DNMT1, DNMT3a and DNMT3b) in healthy subjects and CLL patients as assessed by microarray analysis. A significantly decreased absolute median RNA expression level in CLLs vs. healthy donors was found for DNMT1 (p = 0.0084).
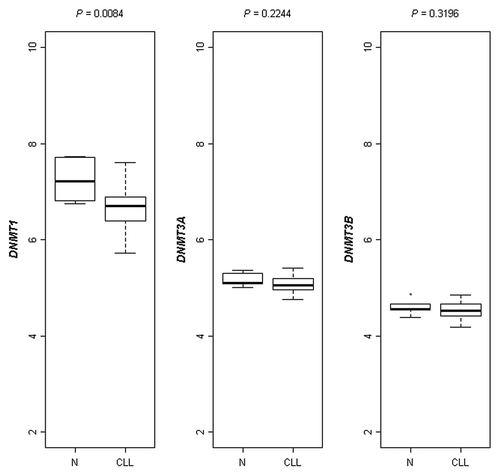
Figure 3. Cox-derived estimated curves according to the SAT-α methylation levels. The curves show the proportional hazard ratio estimate according to SAT-α methylation level below (n = 13, group A) or above (n = 47, group B) the cut-off value (69.0%5mC).