Figures & data
Figure 1 Representative results of MSP analysis of the methylated form of 12 tumor-related genes. M, markers; N, negative control (water blank); P, positive control (universal methylated DNA); T, tumor samples. Samples were scored as methylated when there was a clearly visible band with the methylated primers.
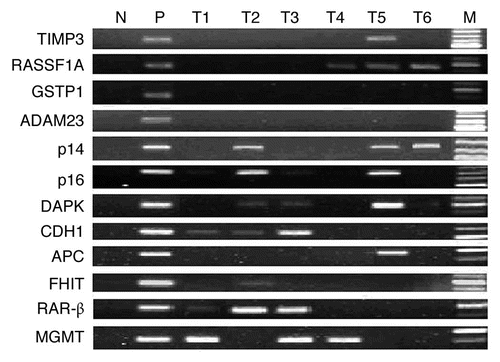
Figure 2 mRNA expression of DNMTs in patients with ESCC (n = 46). (A) Real-time reverse transcription-PCR results for DNMTs. The black bars correspond to non-tumor tissues (NT), and the white bars correspond to ESCC. The means of expression levels of DNMTs are depicted relative to ACTB housekeeping gene expression. Asterisk indicates statistical significance after multiple testing (p < 0.017). (B) DNMT1, 3a and 3b mRNA were expressed in a coordinate manner in the same tumor tissues. “+” indicates elevated mRNA expression as opposed to “−” which indicates a not elevated result. Numbers above the bars indicate the percentage in the total concordant group (+/+ and −/−) and non-concordant group (+/− and −/+). For the association between categories, p = 0.051 for DNMT1/3b, p < 0.001 for DNMT1/3a and p = 0.008 for DNMT3a/3b.
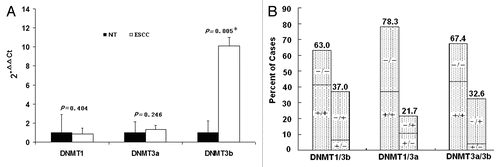
Figure 3 Representative immunohistochemical staining results for DNMT3b protein in two patients with ESCC. Strong nuclear expression was seen in the layer of epithelium in ESCC tissues (A and C). Negative staining was observed in paired non-tumor tissues (B and D). (Original magnification ×400 in A–D).
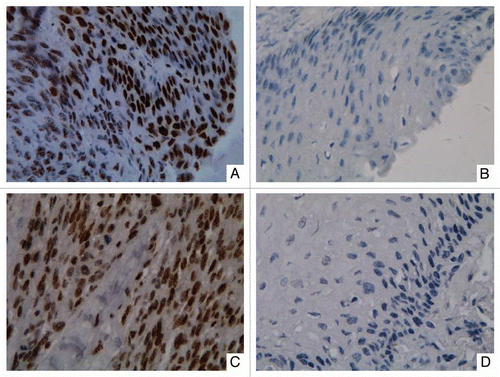
Figure 4 Hypermethylation of cell-free serum DNA at five gene promoter regions in ESCC patients (n = 45) and healthy individuals (n = 15). S, ESCC samples; HI, healthy individuals samples; WBC DNA isolated from the WBCs of healthy volunteers served as a negative control and SssI treated fully methylated DNA served as positive control. NIM was color-scaled between white (representing no methylation detected) and black (indicating that ≥10% of bisulfite-converted input copies were methylated). Asterisk indicates NIM values less than 0.5% but greater than 0.
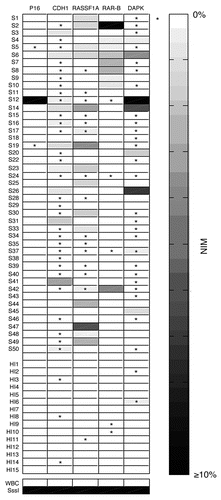
Table 1 Frequency of promoter methylation for individual gene in ESCC patients (n = 47)
Table 2 Correlation between DNA hypermethylation and clinico-pathological parameters of ESCC patients (n = 47)
Table 3 Nuclear immunoreactivity of DNMT3b in ESCC patients (n = 44)
Table 4 Correlation between DNA hypermethylation and mRNA expression of DNMT3b in ESCC (n = 46)
Table 5 Diagnostic information of serum DNA methylation at five genes