Figures & data
Figure 1 Immunocytochemistry of cells isolated from placenta. (A) Cytotrophoblasts stained positively for cytokeratin-7 and (B) did not stain for vimentin-9. The fibroblast fraction stained positively for both (C) cytokeratin-7 and (D) vimentin-9.
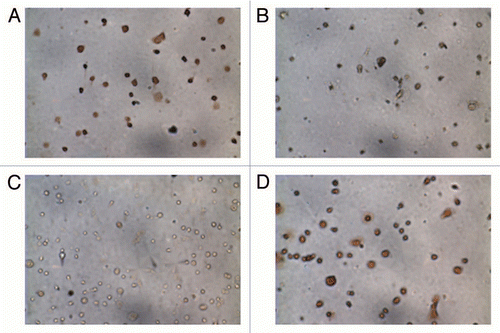
Figure 2 Cluster analysis for placenta, isolated cytotrophoblasts and fibroblasts. Non-hierarchical Euclidean cluster analysis was performed based on gestational age [Column A: 14–16 weeks (early); 17–18 weeks (late)], type of sample [Column B: fibroblast (Fibro), cytotrophoblast (Cyto) and whole placenta (Plac)] and sample number (Column C). The degree of similarity between two samples is given by the sum of the length of the horizontal lines between the samples. We chose the fourth branching to define the clusters (marked by the thick vertical line). There is no clustering by gestational age or sample. The sample type analysis reveals two main clusters: one composed of only cytotrophoblasts and placenta and the other contains mainly fibroblasts. The other three clusters contains only one sample each and may be due to the reduced number of significantly detected probes (24–25,000 vs. > 27,000) for these samples.
![Figure 2 Cluster analysis for placenta, isolated cytotrophoblasts and fibroblasts. Non-hierarchical Euclidean cluster analysis was performed based on gestational age [Column A: 14–16 weeks (early); 17–18 weeks (late)], type of sample [Column B: fibroblast (Fibro), cytotrophoblast (Cyto) and whole placenta (Plac)] and sample number (Column C). The degree of similarity between two samples is given by the sum of the length of the horizontal lines between the samples. We chose the fourth branching to define the clusters (marked by the thick vertical line). There is no clustering by gestational age or sample. The sample type analysis reveals two main clusters: one composed of only cytotrophoblasts and placenta and the other contains mainly fibroblasts. The other three clusters contains only one sample each and may be due to the reduced number of significantly detected probes (24–25,000 vs. > 27,000) for these samples.](/cms/asset/83eadad8-fc18-4fb9-8f1e-be4155629b68/kepi_a_10914196_f0002.gif)
Figure 3 Selection criteria of Differentially Methylated CpG Dinucleotides. (A) The distribution of DNA methylation differences is narrow between cytotrophoblasts and fibroblasts demonstrating that large methylation differences between the two cell types are rare. (B) The scatter plot shows the distribution of differences in methylation as a differential score where a value of >13 or <-13 is equal to a p value of <0.05. The probes with a significant difference in methylation higher than 20% are shown in black. Positive differences (higher cytotrophoblast methylation) in the left upper quadrant and negative differences (higher fibroblast methylation) in the right lower quadrant.
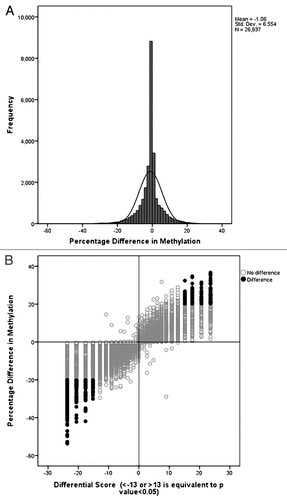
Figure 4 DNA methylation values of the CGB genes in cytotrophoblasts and fibroblasts. (A) CGB genes map to chromosome 19q (middle). At each CpG site there is lower methylation in cytotrophoblasts than in fibroblasts (top). The chromosome band containing the CGB genes is magnified (UCSC genome browser build 36.1 annotation) to illustrate the CpG islands and CpG probes as they relate to the genes (bottom). Six of eight probes show a reduction in methylation, five of these reach our stringent cut-off criteria: CGB3 (probe 1), CGB1, CGB5 and CGB8 (probes 5–8). (B) Box-plot of the methylation values of CpG sites mapping to the CGB genes. Each of the eight CpG probes mapping to CGB genes are represented. The methylation level is depicted on the y-axis. The x-axis shows the CGB gene probes in the order in which they appear in the genome (proximal to distal) but is not to scale. Some genes have two probes and are separated by the interrupted vertical lines. The solid horizontal arrows represent the TSS and point in the direction of transcription. Two CpG probes show no difference in methylation (white box) between the cytotrophoblasts (left) and fibroblasts (right). Probes are less methylated in cytotrophoblasts (left) then in fibroblasts (right). Here each of the array probes mapping to the region are represented by a boxplot. The statistically significant different probes (Wilcoxon rank-sum test, p < 0.05) reaching or passing the 20% cut-off are represented by a dark gray bar while the clear gray bar corresponds to the probe that has <20% difference in methylation but remains statistically significant.
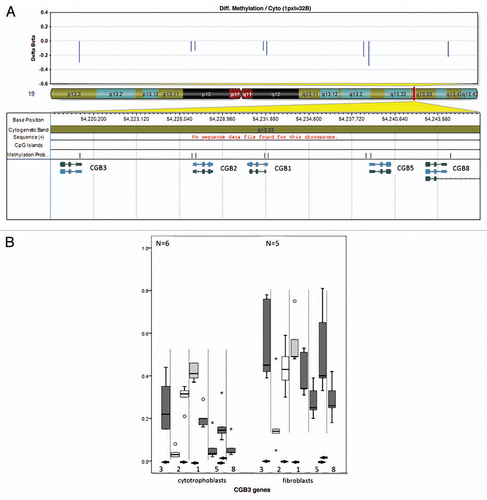
Figure 5 DNA methylation values of CpG sites mapping to tumor suppressor genes in cytotrophoblasts and fibroblasts. For each of the box plots the y-axis corresponds to methylation levels and the x-axis are the probes; the solid horizontal arrows indicate the direction of transcription. All the CpG sites are between 2,000 bp upstream and 500 bp downstream of the TSS (start point of the arrows). For each gene all the probes corresponding to a CpG Island that has at least one differentially methylated probe between cytotrophoblasts and fibroblasts are shown. Only dark grey boxes correspond to statistical significantly different probes (Wilcoxon rank-sum test, p < 0.05). (A) The solid vertical lines on the APC gene graph separate probes of two consecutive but distinct CpG islands. Of the 6 APC probes, one shows a non-statistically significant difference higher than 20% (light grey box). (B) DAP2IP, (C) PRKCDBP and (D) RASSF5 have multiple probes that show no difference in methylation (white boxes). (E) TP73 shows a non-statistically significant difference higher than 20% (light grey box). (F) All 3 probes corresponding to RASSF1 are statistically different (dark grey box).
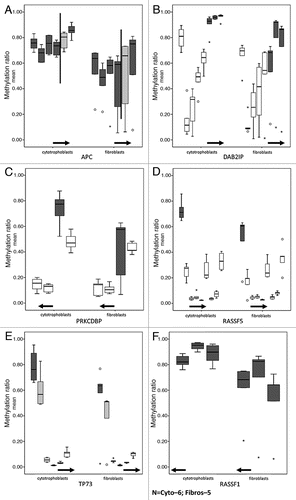
Figure 6 Pyrosequencing validation of DNA methylation measured by microarrays. Three CpG sites mapping to the promoter region of (A) CGB5 and the tumor suppressor genes (B) TP73 and (C) APC were analyzed. For each gene three box plots are shown. From left to right: the first corresponds to the values for each CpG site as measured in the pyrosequencing assay, the second to the average of the values of all the CpG sites for each pyrosequencing assay and the third plot corresponds to the methylation value measured by the array. The white boxes correspond to non-statistically significantly different CpG sites and the grey boxes to statistically significantly different CpG sites between cytotrophoblast and fibroblast samples. The light grey boxes show the CpG of the pyrosequencing target that corresponds to the one that was targeted by the array.
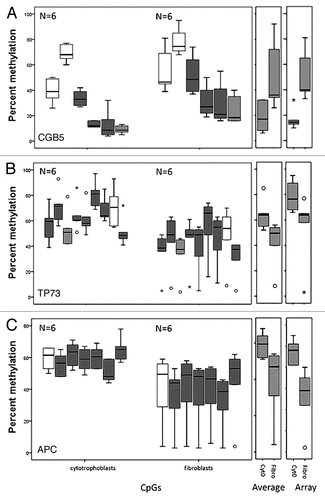
Table 1 Probes more methylated in cytotrophoblasts than fibroblasts mapping to tumor suppressor genes