Figures & data
Figure 1 (A) Dlk1-Gtl2 imprinting cluster, including transcriptional start sites (arrows) and transcription units (hatched boxes). (B) Portion of IG-DMR analyzed in this study. The 458 bp region analyzed by bisulfite mutagenesis and DNA sequencing corresponds to positions 110,766,298-110,766,755, NC_000078.5 (black box). Polymorphisms (*) between C57BL/6J and Mus musculus castaneus are as follows: (B6/CAST): 110,766,439 (A/G), 110,776,579 (G/A), 110,766,774 (G/A), 110,766,902-110,766,904 (TTT/TT), 110,767,052 (A/G). (C) Schematic of the Gtl2-DMR, including the Gtl2 transcriptional start site (arrow) and exon 1 (hatched box); +1 corresponds to position 110,779,206. Regions analyzed correspond to positions 110,778,378-110,778,966 and 110,779,331-110,780,052. Polymorphisms are as follows: 110,779,741 (G/A), 110,779,881 (A/G), 110,780,030-110,780,031 (AA/GC).
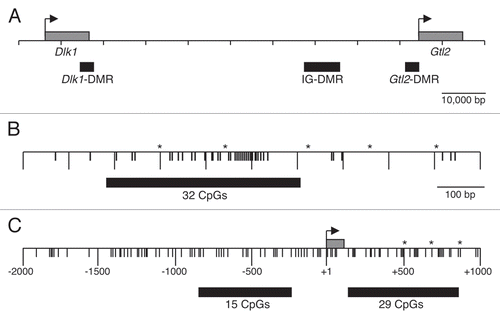
Figure 2 Paternal allele-specific methylation of the IG-DMR is inherited from sperm. Bisulfite mutagenesis and sequencing of DNA from B6 × CAST and CAST × B6 F1 hybrid liver and B6 × CAST F1 hybrid spermatozoa. Each circle represents one of 32 potentially methylated CpG dinucleotides, the first one located at position 110,766,345 (NC_000078.5). Each row of circles represents an individual strand sequenced. Filled circles represent methylated cytosines, open circles represent unmethylated cytosines, absent circles represent positions at which methylation data was not obtained.
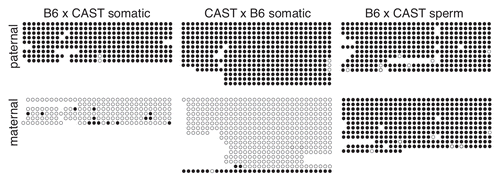
Figure 3 Methylation of the paternal IG-DMR is maintained during pre- and post-implantation development. Bisulfite mutagenesis and sequencing of DNA from B6 × CAST F1 hybrid embryos. Details as described in .
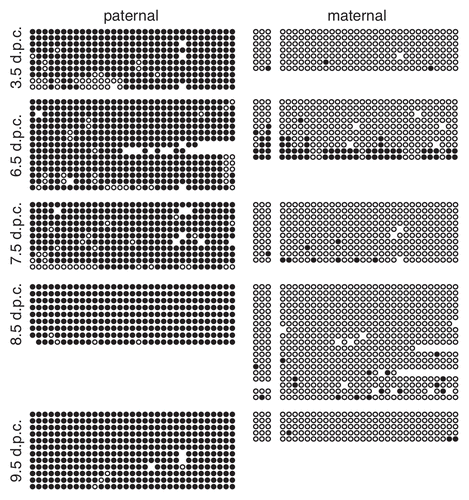
Figure 4 Paternal allele-specific methylation of the Gtl2-DMR is not inherited from sperm. Each circle represents one of 29 potentially methylated CpG dinucleotides, the first one located at position 110,779,349 (NC_000078.5). Details as described in .
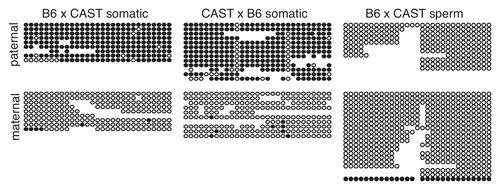
Figure 5 Methylation of the paternal Gtl2-DMR is acquired between 3.5 and 6.5 d.p.c. Bisulfite analysis and sequencing of DNA from B6 × CAST F1 hybrid embryos. Details as described in and .
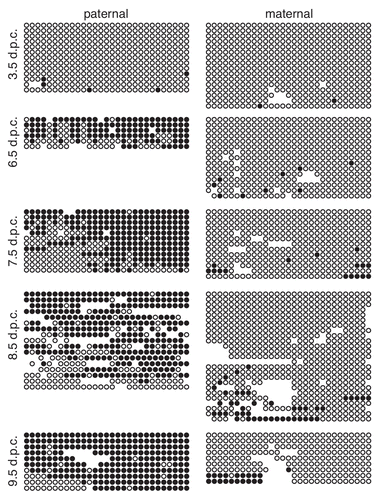
Figure 6 Expression analysis of Gtl2 in embryonic tissues. (A) Genomic structure of Gtl2, including exons (black boxes), B6 vs. CAST polymorphisms (*), major transcripts, primers used for RT-PCR and CpG-rich regions analyzed in this study (grey boxes). (B and C) RT-PCR of RNA from neonatal B6 liver and 6.5–9.5 d.p.c. B6 × CAST embryos using PCR primers A/B (B) and F/R (C). MW, molecular weight marker; 0, no template; + and − represent samples with and without reverse transcriptase (RT). (D) RT-PCR products from (B and C) were digested with SfcI and NgoMIV, respectively, to distinguish between products derived from maternal B6 vs. paternal CAST alleles. (E) RT-PCR of RNA from B6 brain, CAST brain and 3.5 d.p.c. B6 × CAST blastocysts using PCR primers F/R; B-N, C-N and 3.5 d.p.c. products were generated using a nested PCR approach described in the Materials and Methods. Alternative splice forms were detected in B6 and CAST samples; only the larger splice variant was detected in the +RT 3.5 d.p.c. sample. (F) RT-PCR products from E were digested with NgoMIV to distinguish between products derived from digested B6 vs. undigested CAST alleles.
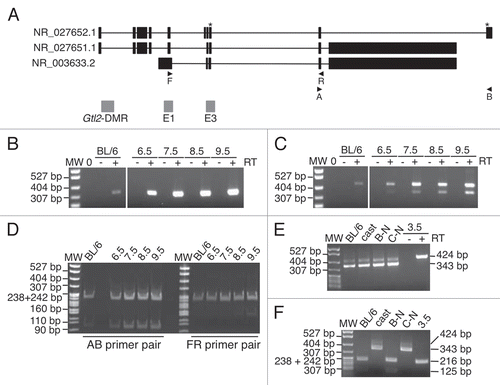
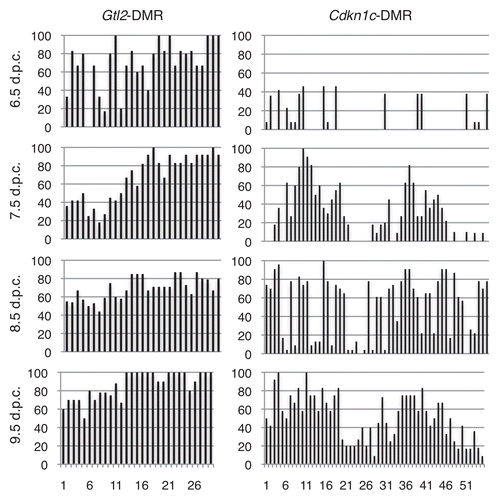
Figure 7 Methylation is acquired at the Gtl2 and Cdkn1c DMRs during different developmental stages. Graphs summarizing the percentage of methylation on paternal strands at each CpG dinucleotide at the Gtl2- and Cdkn1c-DMRs in 6.5–9.5 d.p.c. embryos. Cdkn1c data derived from Bhogal et al.Citation14