Figures & data
Figure 1. Early NE specification is not affected by DNMT3B knockdown. (A) protein gel blot analyses of DNMT3B to confirm DNMT3B knockdown efficiency in hESCs. β-actin was used as a loading control. For each lane, the DNMT3B/ β-actin ratio is shown, as determined by quantification of the bands. The relative amount of remaining DNMT3B in the KDs was determined relative to undifferentiated H9 hESCs. (B) Schematic of NE differentiation protocol. Morphogens can be added during differentiation as follows: * = Addition of exogenous morphogens, such as retinoic acid, at day 10 (d10) of differentiation is required for robust expression of neural patterning genes around d17. ** = Addition of exogenous morphogens to hESC-derived rosettes beginning at d16 is required for robust expression of NC patterning genes at d28. (C) Phase contrast images for NE at d10. (D) Immunocytochemistry for PAX6 and TUJ1 at d10. Cell nuclei were counterstained with DAPI.
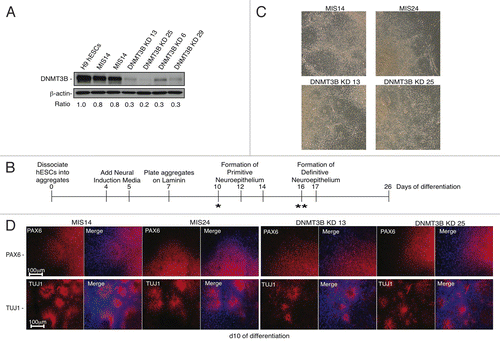
Figure 3. DNMT3B knockdown enhances NC lineage differentiation and specification. (A, B) Gene expression analyses of PAX7 (A), or NGFR (B) at d5 and d7 of differentiation. Data was normalized to undifferentiated H9 hESCs and displayed as a Log2 ratio. (C) Immunocytochemistry for PAX7 and NGFR at d8. Cell nuclei were counterstained with DAPI. (D, E) Gene expression analyses of early NC specifiers at d10 (D), or NC lineage fate determinants (E) at d12. Gene expression was also quantified for the NC lineage fate determinants ASCL1, PHOX2B, HOXC8, and S100β at d12 of differentiation. Data was normalized to undifferentiated H9 hESCs and displayed as a Log2 ratio. The p values displayed correspond as follows: * < 0.10, ** < 0.05, *** < 0.01.
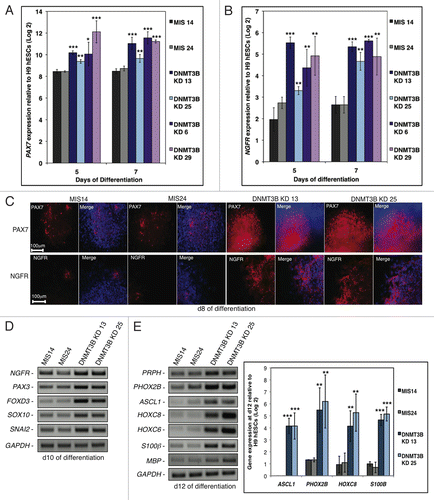
Figure 2.Expression of neural genes that specify regional identity is altered when DNMT3B is knocked down. (A) Gene expression analyses of NE regional specifiers at d12 of differentiation. Brain regions specified by these genes are indicated. (B) Immunocytochemistry for HOXB4 and ISL1 at d12. Cell nuclei were counterstained with DAPI. (C) Gene expression quantification of EN1, TH, HOXB4, ISL1 at d0 (top) and d12 (bottom) of differentiation. Data was normalized to undifferentiated H9 hESCs and displayed as a Log2 ratio. The p values displayed correspond as follows: * = < 0.05.
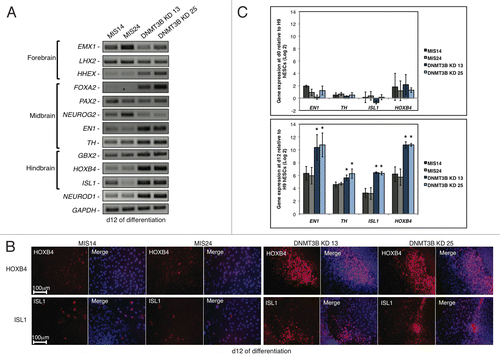
Table 1. DNA methylation levels at pericentromeric chromosomal regions
Figure 4. Aberrant DNA methylation in DNMT3B KDs. (A) Chromosome-wide distribution of DNA methylation along X chromosome. HMDs with > 80% DNA methylation (common to all samples) are marked in blue. (B) The average percent methylation observed within HMDs and PMDs along X chromosome. Control samples used include male H1 hESCs and male cerebral cortex, which showed no differences in the methylation of these two domains. (C) The average percent methylation of autosomal domains. Control sample used include male H1 hESCs. The p values displayed correspond as follows: *** = < 0.001.
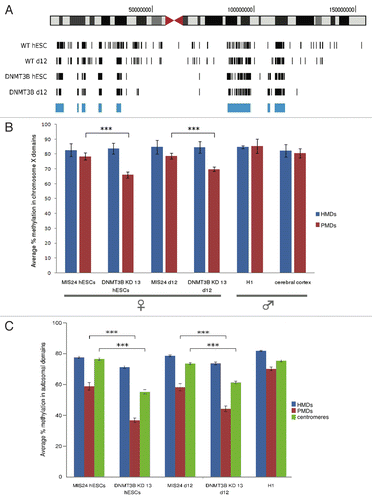
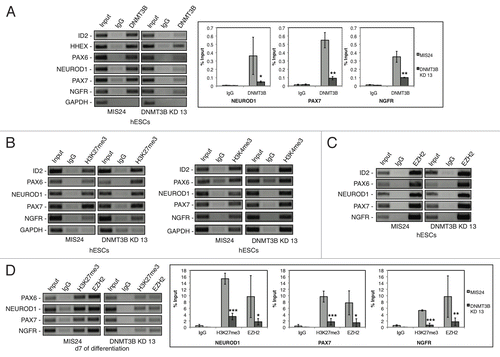
Figure 5. DNMT3B associates with the promoters of early neural determinant genes. (A) DNMT3B association at PAX6, NEUORD1, PAX7, and NGFR promoters in MIS24 and DNMT3B KD 13 hESCs. DNMT3B was also enriched at known target promoters of ID2 and HHEX, but not at GAPDH. Quantification of ChIP was performed and displayed as % input for each sample. (B) Promoter regions are “bivalent” in hESCs, with H3K27me3 and H3K4me3. (C) Association of EZH2 at the promoters in hESCs. (D) Levels of H3K27me3 and EZH2 at d7 of differentiation. Quantification of ChIP was performed and displayed as % input for each sample. The p values displayed correspond as follows: * = < 0.08, ** = < 0.05, *** = < 0.01.