Figures & data
Figure 1.sr45–1 de novo DNA methylation phenotype. (A) Flowering time of Columbia, sr45–1, dcl3–1 and sr45–1, dcl3–1 double before and after FWA transformation. Flowering time is measured as the total number of leaves at the time of flowering. (B) Methylation levels at endogenous and transgenic FWA after FWA transformation. The 594 base-pair repeated region in the 5′ UTR was analyzed. The methylation state of the FWA endogene should remain unaltered by the presence of the FWA transgene. All samples were analyzed in the T1 generation.
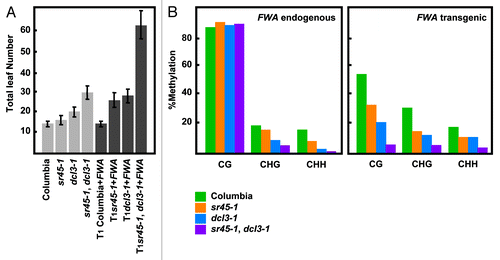
Figure 2. Analysis of the FWA transgene methylation generationally. Methylation levels at endogenous and transgenic FWA after FWA transformation. Endogenous FWA was only analyzed in the T1 generation. Transgenic FWA was analyzed during three generations after transformation.
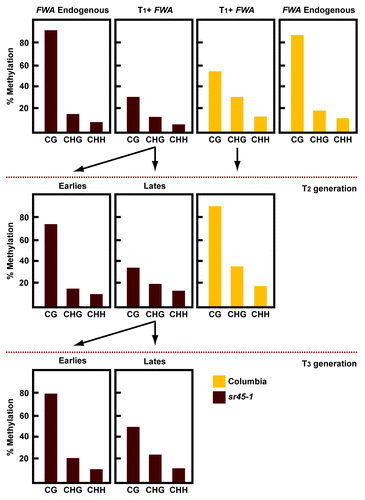
Figure 3.sr45–1 maintenance DNA methylation phenotype. (A) Sodium bisulfite analysis of an 180 base-pair region of the MEA-ISR locus. (B) AtSN1 Chop-qPCR assay. Genomic DNA was digested with the methylation sensitive enzyme HaeIII, which recognizes three sites in AtSN1. Amplification of AtSN1 was quantified by Real Time PCR, and signal was normalized to undigested DNA. HaeIII, is blocked by C methylation in GGCC context. (C) MEA-ISR DNA gel blot. MspI digested genomic DNA was probed with MEA-ISR. (D) REP12 and Ta3 DNA gel blot. MspI digested genomic DNA was probed with REP12 or Ta3. MspI is blocked by methylation of the external C in CCGG context. (E) RT-PCR showing expression levels of REP12 and Ta3. UBQ10 expression is showed as a loading control.
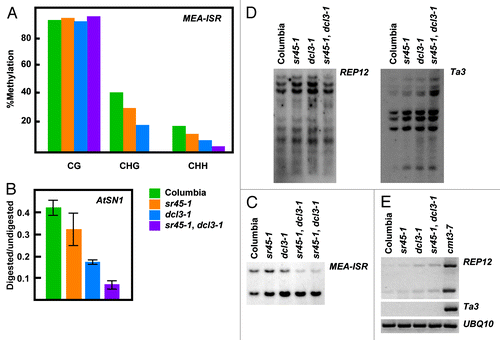
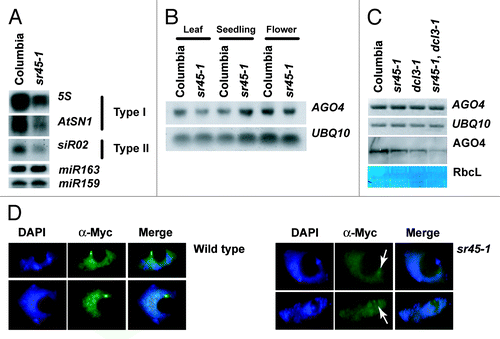
Figure 4. Placement of SR45 in RdDM pathway. (A) RNA gel blots showing siRNAs abundance at both type I and II loci. Hybridization with miR163 is shown as a loading control for 5S siRNAs and hybridization with miR159 is shown as a loading control for AtSN1 and siR02. (B) RNA gel blot showing AGO4 expression in leaves, seedlings, and flowers. Hybridization with UBQ10 is shown as a loading control. (C) RT-PCR and protein gel blot showing the expression and abundance of AGO4/AGO4. UBQ10 expression is showed as a loading control for RT-PCR and amido black staining of the RUBISCO large subunit is shown as a loading control for the protein gel blot. (D) Immunofluorescent microscopy showing AGO4 localization in 4xmyc::AGO4 (Columbia) and 4xmyc::AGO4 (sr45–1) backgrounds. White arrows indicate the position of AGO4 in the sr45–1 panel.