Figures & data
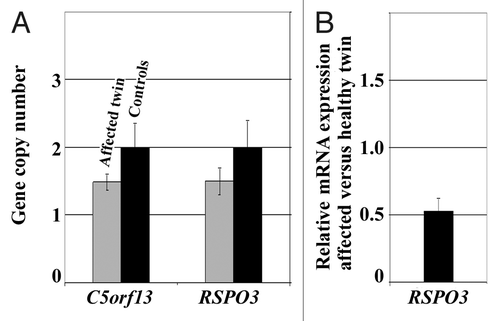
Figure 1. Box plots showing the distribution of BRCA1 methylation values in fibroblasts of 10 one-cancer and 10 two-cancer patients. The star symbol (extreme outlier) represents the affected twin. The median is represented by a horizontal line. The bottom of the box indicates the 25th percentile, the top the 75th percentile. The T bars extend from the boxes to 1.5 times the height of the box.
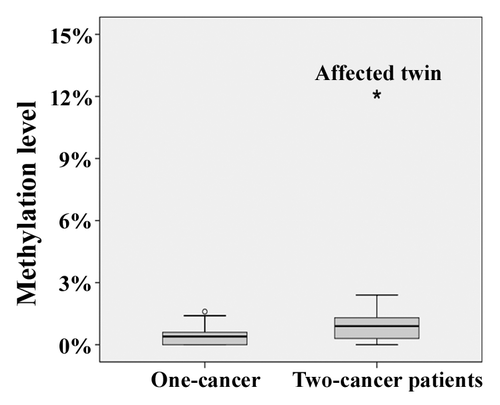
Figure 2. Methylation patterns of the BRCA1 promoter in fibroblasts of the affected twin and her healthy sister. Each line represents an individual allele (DNA molecule) analyzed by bisulfite plasmid sequencing. Filled circles indicate methylated CpG and open circles unmethylated CpG sites. Missing dots indicate CpG sites that could not be analyzed because of poor sequence quality. The five framed CpG sites correspond to those in the bisulfite pyrosequencing assay. Both twin sisters display alleles with single CpG (stochastic) methylation errors. Six of 47 analyzed alleles (indicated by arrows) in the affected twin represent epimutations with the majority of CpGs being aberrantly methylated, whereas all 47 alleles in the healthy twin are hypomethylated.
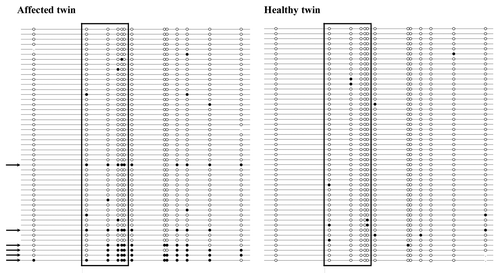
Figure 3. (A) Constitutive protein expression of ACTB (control), ATM, BRCA1, BRCA2, MLH1, RAD51, and TP53 in untreated fibroblasts of the affected vs. the healthy twin. Relative protein expression (z ratio normalized by log10 transformation and z scores) was measured by antibody microarrays using three biological replicates. Note the decreased BRCA1 level (75% expression ratio) in the affected twin. (B) Differential induction of BRCA1 protein in fibroblasts of the healthy twin (gray bars) and the affected twin (black bars) at 1 h and 4 h after 1 Gy γ-irradiation. Relative protein expression in treated vs. untreated cells of each twin was measured by antibody microarrays using three biological replicates.
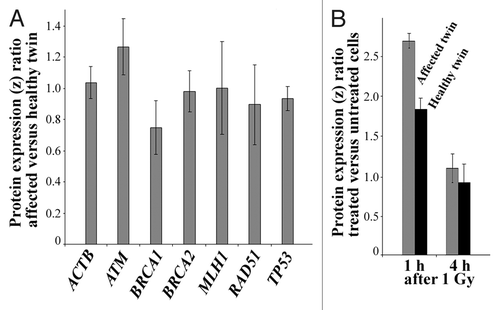
Figure 4. (A) C5orf13 and RSPO3 copy numbers in fibroblast cells of the two-cancer twin, compared with healthy controls, as determined by qPCR (using the RFC3 gene copy number as reference). Standard deviations represent three independent DNA samples (biological replicates) of the affected twin and a mixture of control DNAs. (B) RSPO3 mRNA expression ratio between the affected twin and her healthy sister, as determined by GeneAtlas microarrays. Standard deviations represent RNA samples from four different cell cultures of the affected and the healthy twin, respectively.