Figures & data
Figure 1. Cellular dynamics and signaling in growing ommatidial clusters. (A) The growth and maturation of the ommatidium, from the precluster stage to the specification of the cone cells. (i) Shows the five precluster cells surrounded by a sea of unspecified cells. (ii) Cells are systematically recruited to the unit with the first three (white shapes) incorporated along the R2/8/5 face. (iii) Two of the three begin to differentiate (R1/6 – dark gray), while the cell between them (R7 – white shape) does not. (iv) Cells are next added to flanking positions of the cluster (white shapes) as R7 begins differentiation. (v) Two more cone cell precursors are added above and below the cluster (white shapes). (vi) All seven of the newly added cells of the cluster differentiate. (B) A model for why R1/6 activate N strongly in the R7 precursor. (i) Spitz (gray arrow) is released from the R2/5 precluster cells. The R1/6 precursors directly about these cells and (ii) are rapidly specified (dark gray), and express Dl (black line shapes). The Dl expression activates N in the R7 precursor. (iii) When the flanking cone cells join the unit, they receive a potent N activation that prevents them from responding to Spitz. (C) Schematic depiction of Sev expression and activation. (i) As R7 and the flanking cone cells differentiate, they too express high levels of Dl ensuring that all four cone precurors (and R7) experience high N activation. (ii) N transcriptionally activates sev in these cells leading to a high level of Sev (speckling) in these cells. (iii) The ligand for Sev is Boss (thick black line) that is exclusively expressed on membrane of R8. The only cell expressing Sev and contacting R8 is the R7 precursor. This cell now selectively experiences high-level RTK activation (*). (D) Summary diagram depicting the fate of the photoreceptor precursor cells in response to differential activation of the N and RTK pathways. R1/6 cells are specified when DER signaling is active and there is low N activity. R7 cells are specified when N activity is high and both DER and Sev RTK pathways are engaged. The cone cells are specified when N activity is high but only the DER RTK pathway is active. Even though the cone cells express Sev, they do not contact the Boss expressing R8 cell, so the Sev RTK pathway is not engaged.
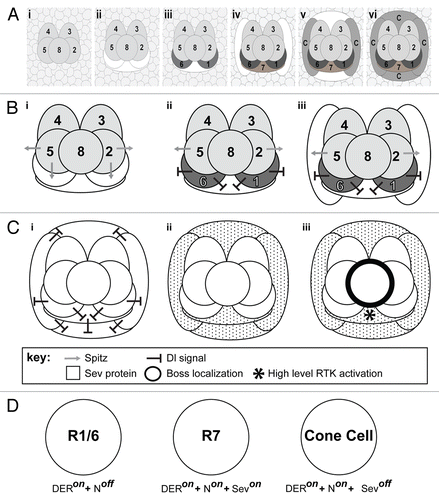
Figure 2. Details of the RTK transduction pathway. (A) In the absence of ligands the RTKs (Sev and DER) are inactive. The phyllopod gene is not transcribed and Ttk acts as a transcriptional repressor in the cells, and the photoreceptor fate is blocked. (B) In the presence of ligand, RTK transduction occurs via Ras leading to MAPK phosphorylation and translocation to the nucleus where it promotes phyl transcription. The resulting Phyl protein recruits the Sina E3-ubiquitin ligase to poly-ubiquitinate Ttk,Citation16,Citation17 thereby targeting it for degradation, and releasing the block on photoreceptor differentiation.
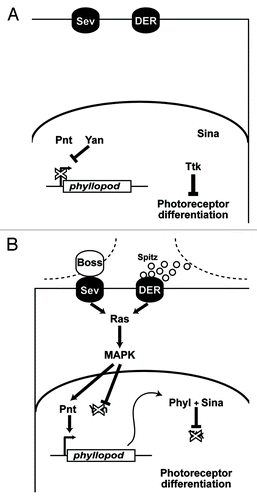
Figure 3. Structural similarities between the organization of the five photoreceptor cluster of the Phyllopoda and the precluster of Drosophila. (A) Shows a drawing of the five photoreceptors of adult Leptodora kindtii adapted from.Citation24 The image has been rotated from the original, and the numbers of the photoreceptors changed to correspond with the Drosophila nomenclature. The hatching in the middle represents the rhadom of the photoreceptors. (B) Shows a TEM photograph through the Drosophila precluster. Although the two images are from different developmental stages, and the R8 cells are of dramatically different sizes, the overall topology of the cells is largely conserved.
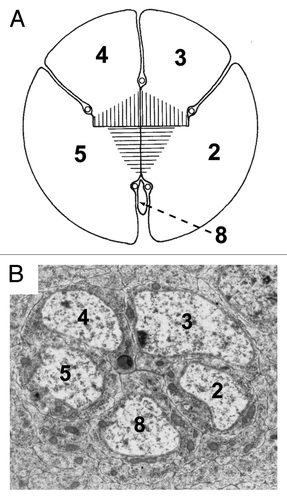
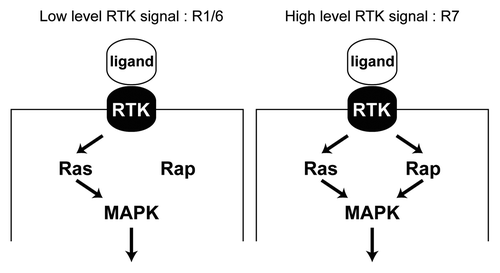
Figure 4. Differential requirement for Rap in RTK transduction. (A) In the R1/6 precursors a mild RTK signal is transduced, where Ras alone suffices for this purpose. Rap is likely engaged, but is not required since its removal does not compromise the specification of these cells.Citation1 (B) In the R7 precursor a robust RTK signal is transduced and this requires the combined presence of both Ras and Rap. Removal of either prevents adequate transduction of the pathway and a failure of the R7 to be specified.