Figures & data
Figure 1. The actin-bundling protein Forked is required during tracheal system development. (A–D) Percentage of embryos with the indicated number (in different colors) of dorsal branches (DBs) misguided (A), unfused DBs (B), terminal intracellular lumen misguided or bifurcated (C), or with extra DSRF-terminal cells (D). n is the total number of fixed embryos analyzed. Note that f mutant embryos (f36 FBal0003950) display stronger defects than sn mutants (snP1. FBal0035641) in all aspects analyzed. ***P < 0,001 analyzed by Fisher's exact test (http://www.langsrud.com/fisher.htm#INTRO). (E–G) Confocal projections of fixed control (yw) (E), snP1 (F) and f36a (G) embryos at the end of embryogenesis stained with the luminal marker 2A12 (red) that labels the tracheal tree and the terminal cell marker DSRF (white). (H) Close-up of two DBs of a f36a; btlGal4 UAS-srcGFP mutant labeled with 2A12, DSRF and GFP (which highlights cell morphology due to the membrane marker Src fused to GFP under the control of the breathless (btl)-Gal4 tracheal driver). (I–L) Confocal projections of fixed sn3f36a embryos (FBst0306268) stained with 2A12 (red) and DSRF (white). (K–L) are close-ups of two DBs of different embryos. White arrows point to the presumptive point of fusion between contralateral DBs at the dorsal midline. Blue arrows point to the intracellular terminal lumen of terminal branches. Note that terminal lumina correctly extends ventrally in control embryos (E), but often turn dorsally (F and H) or hardly extend (J and L) in mutant conditions. Purple arrows point to the individual (E), extra (F-H) or missing (I-L) DSRF-expressing cells of DB in different conditions. Yellow asterisks (I) indicate missing DBs. In all cases yw was used as the control. Scale Bar: 15 μm.
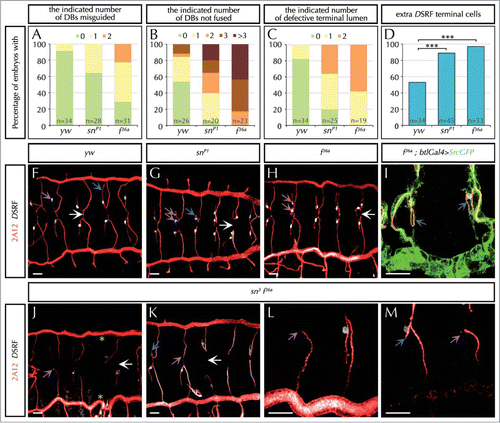
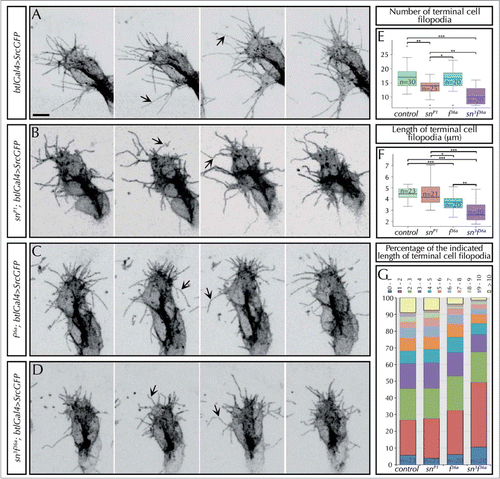
Figure 2. Singed and Forked control the number, length and stiffness of tracheal filopodia. (A–D) Stills from in-vivo movies taken approximately every 10 s of embryos carrying btlGal4 > SrcGFP (which allows to clearly visualize the morphology and dynamics of filopodia) in an otherwise wild-type (A), snP1 (B), f36a (C) or sn3f36a (D) backgrounds. The images show the tip of DBs of stage 14–15 embryos. Arrows point to straight (in control, A) or curved filopodia (in sn, f or snf mutants). Note also the presence of conspicuously shorter filopodia in f or snf mutants. Scale Bar: 5 μm. (E) Quantification of the number of filopodia in the terminal cells of DBs of fixed embryos at stage 16 carrying btlGal4 > SrcGFP in a control or, snP1, f36a and sn3f36a mutant background. Note the decrease in the number of filopodia in sn mutants, as well as sn f double mutants. *P < 0,05; **P < 0,01; ***P < 0,001 were obtained using two-tail t test of R-Commander software. n = number of terminal cells analyzed. (F) Quantification of the length (in microns) of filopodia in the terminal cells of DBs of fixed embryos at stage 16 carrying btlGal4 > SrcGFP in a control or, snP1, f36a and sn3f36a mutant background. f terminal cell filopodia are significantly shorter than control and sn mutant ones. sn f terminal cell filopodia are much shorter than the previous cases. *P < 0,05; **P < 0,01; ***P < 0,001 were obtained using two-tail t test of R-Commander software. n = number of terminal cells analyzed. (G) Distribution of the length of filopodia of terminal cells (represented in different color/micron) in control, snP1, f36a and sn3f36a fixed embryos at stage 16. Note that in sn f mutants almost 50% of filopodia are shorter than 2 μm and hardly any filopodia extend longer than 10 μm. Typically, long filopodia (> 10 μm) are associated with the ventral cytoplasmic extension that generates the terminal branch. n = number of terminal cells analyzed. In all cases btlGal4 > SrcGFP was used as control.