Figures & data
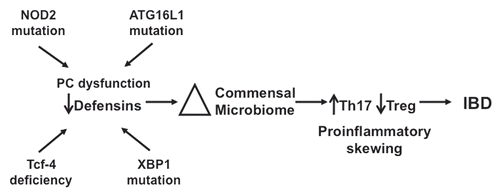
Figure 1 Comparison of microbial communities of distal small intestines from DEFA5 TG and MMP7−/− mice. Genomic DNA was isolated from the distal small intestines of age matched DEFA5 TG mice and their wild type littermates, and from MMP7−/− mice and their wild type littermates. Microbial community analysis was done based on 16S rRNA gene sequence, using high through put Sanger sequencing (A) and quantitative PCR (B). The stacked graphs show the relative percentages of the bacterial groups identified by each method. Percentages were obtained for Sanger subclone sequencing using the total number of sequences obtained as the denominator. Percentages were obtained for qPCR by using total bacterial 16S gene copies as the denominator, determined by amplifying each sample with universal bacterial primers. Figure reproduced from original publication by Salzman et al.Citation32
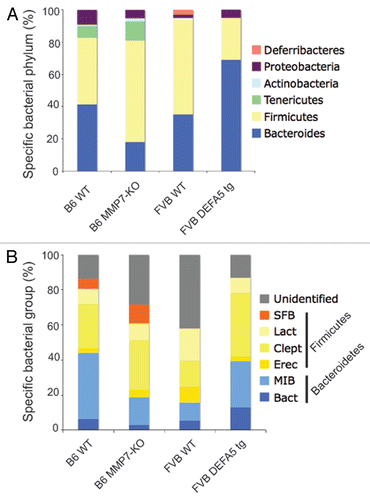
Figure 2 Segmented filamentous bacteria (SFB) is lost in DEFA5 TG mice. Fluorescence in situ hybridization was performed on the terminal 1.5 cm section of distal small intestine from DEFA5 TG and MMP7−/− mice, for the detection of total bacteria and for SFB. Tissue sections were hybridized with a combination of oligonucleotide probes for all bacteria (Texas red labeled) and SFB (6Fam labeled) and examined by fluorescence microscopy. Images were overlaid in Adobe Photoshop. Bacteria that hybridize with the universal bacterial probe fluoresce red. Bacteria that co-hybrizide with the universal bacterial probe and the specific SFB probe appear yellow. In the small intestine, segmented filamentous bacteria (yellow) directly contact the intestinal epithelium (green), unlike the rest of the commensal microbiota (red), which are located in the intestinal mucus. Arrows point to SFB bacteria. Figure is a composite of unpublished and published data by Salzman et al.Citation32
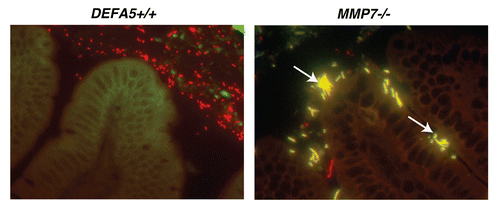
Figure 3 HD5 expression related to NOD2 genotype in patients with ileal CD. HD5 mRNA expression was determined by RT -qPCR of RNA isolated from ileal biopsies from patients with CD and controls. Samples were grouped by NOD2 genotype. All patients with CD had significant reductions in HD5 mRNA expression, irrespective of NOD2 genotype. However, there were significantly greater reductions in HD5 expression in patients with the SNP13 NOD2 mutation. The SNP13 mutation has shown increased association with severe ileal disease.Citation48,Citation49 Composite figure and data from Wehkamp et al.Citation44 redrawn by Dr. Charles Bevins, UC Davis School of Medicine, Davis, CA , Copyright (2005) National Academy of Sciences USA.
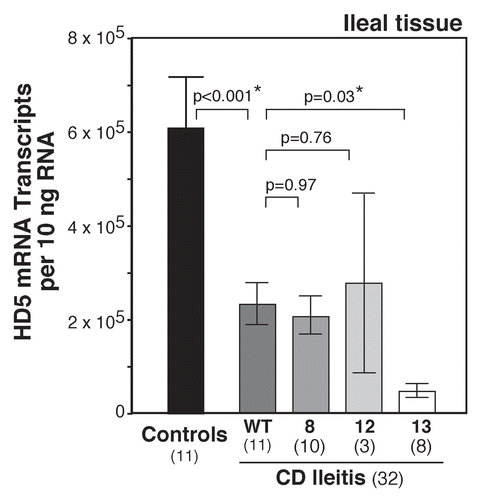
Figure 4 An exploratory model for the role of defensin deficiency in CD. Several genetic mutations (NOD2, AT G16L1, XBP1) lead to either global PC dysfunction or specific defensin deficiency (NOD2, Tcf-4). Reduction in PC defensin expression results in alterations in the intestinal bacterial microbiome. Shifts in the biome lead to induction of Th17 cells and proinflammatory skewing of the Th17:Treg balance, resulting in predisposition to the persistent and chronic inflammation associated with CD.