Figures & data
Figure 1. Weights of mice over the first 364 d post weaning (dpw). For each litter two male mice were co-housed and two female mice were co-housed. In addition, two males and two females from the second litter were each individually housed. The vertical bars along the bottom indicate samples that were used for the microbiome analysis described in the current study.
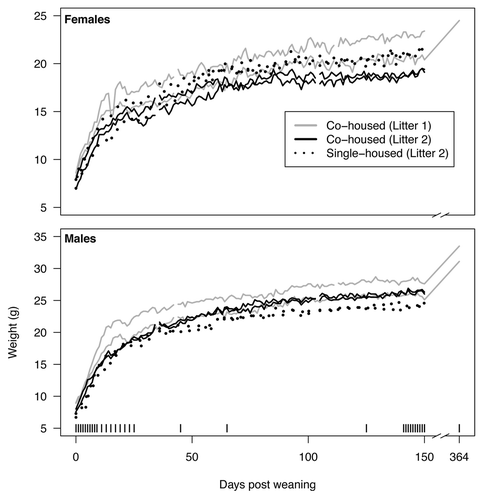
Figure 2. Temporal variation of the community structure of the murine gut microbiome for all 12 mice together as well as those from two female and one male animal over the first 364 dpw as represented in NMDS plots using the ΘYC distance metric. The stress for this representation was 0.15. Samples collected between 0 and 9 dpw are in blue, those from 11 to 140 dpw are in green, those from 141–150 dpw are in red, and samples from 364 dpw are in black. Larger characters are closer in the unrepresented axis and smaller characters are further into the unrepresented axis. Animated versions of the NMDS plots for each animal and all points together are available as supplementary data.
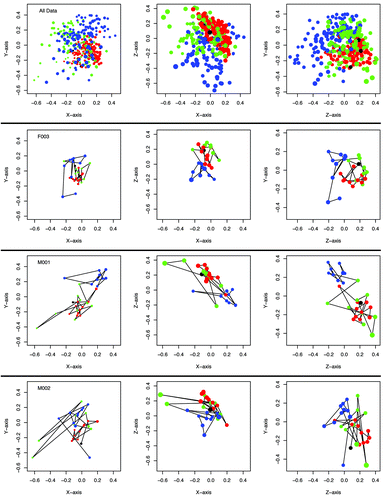
Table 1. The difference in average relative abundance of OTUs between the early and late periods
Figure 3. Average ΘYC distances between days for the same animal (self-comparison) and between different animals (other-comparison) for 0 to 9 dpw (blue) and 141 and 151 (red) dpw. Error bars represent the 95% confidence interval.
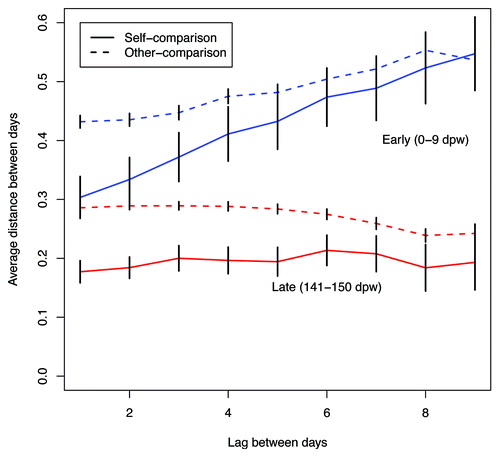
Table 2. The difference between the coefficient of variation for the early and late periods for each OTU
Figure 4. Relative difference in root mean squared θYC distances to the early (0–9 dpw) and late (141–150 dpw) samples for each animal. Rectangles with “NC” indicate that the samples were not characterized and those with “ND” indicate those samples that had fewer than 1,900 sequences.
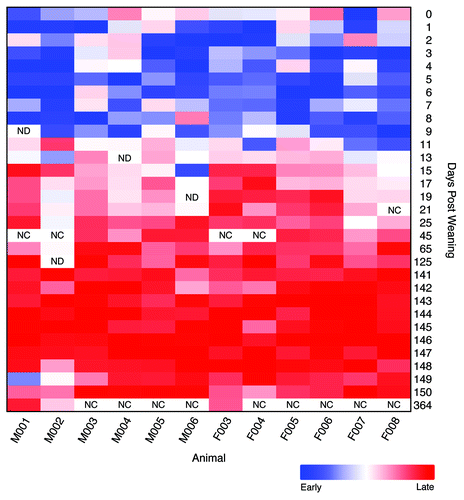
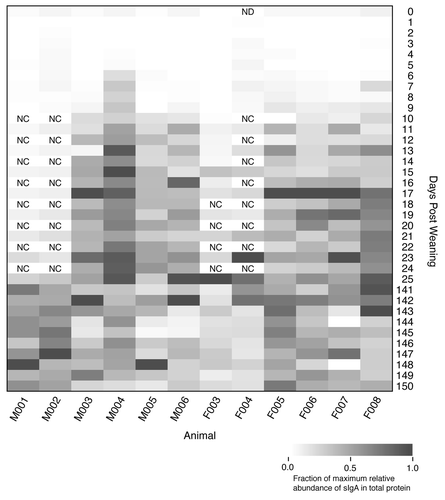
Figure 5. Relative percentage of secretory IgA as total protein in the feces of each animal over the first 150 dpw. Rectangles with “NC” indicate that the samples were not characterized and the sample with “ND” did not yield useable data.