Figures & data
Table 1. Quantitative real time PCR primers for targeted mouse genes
Figure 1. Relative expression of target genes in the ileum of Nod1 (A) and Nod2 (B) littermates (mean control value set to 1).
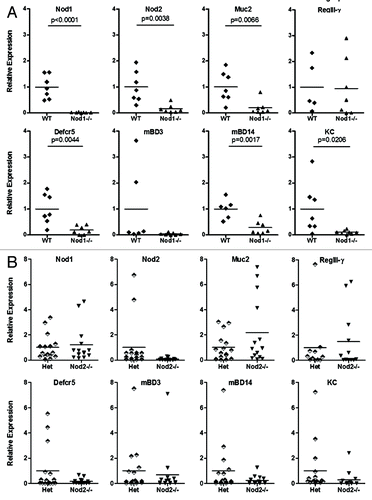
Table 2. Quantitative real time PCR primers for bacterial group-specific16S rDNA
Figure 2. Relative abundance of targeted bacterial groups in the ileum of Nod1 non-littermates, including co-caged WT and Nod1−/− females (A) and Nod1 littermates (B).
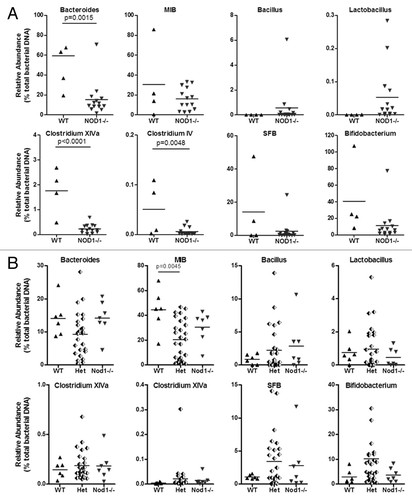
Figure 3. NMS ordination of bacterial community structure in the ileum of WT and Nod1−/− mice under different housing conditions. Littermates tend to cluster closely together while there is greater separation between genotypes of the non-littermate mice. Co-caged mice (open symbols) tend to separate from the other groups.
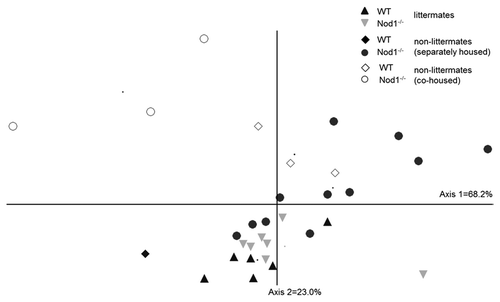
Figure 4. Relative abundance of targeted bacterial groups in the ileum of Het and Nod2−/− littermates. Variation within litters is shown by the different shading of points on each graph.
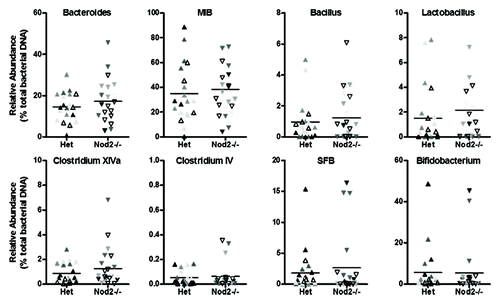
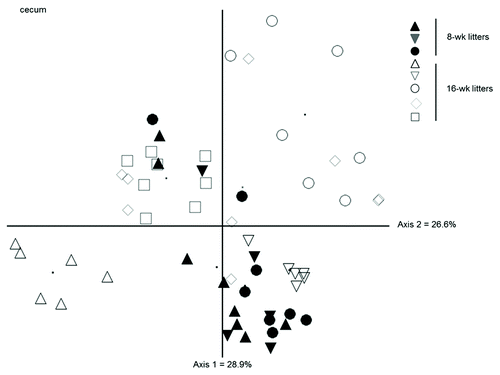
Figure 5. NMS ordination of bacterial community structure (from fragment analysis by LH-PCR) in the cecum of WT and Rip2−/− showing variation between litters at 8 and 16 weeks. The community structure of 8-week mice did not vary significantly from each other but the 16-week mice varied from the 8-week mice as well as with each other, regardless of genotype.