Figures & data
Figure 1. Rhesus macaque IL-12 expression vector. Map of the dual promoter plasmid (AG157) encoding the rmIL-12p70 cytokine. The rmIL-12p40 and rmIL-12 p35 genes are under the control of the human CMV and the simian CMV promoter, respectively. The polyadenylation signal from bovine growth hormone (BGH) and the simian virus 40 (SV40) are used for the p40 and the p35 subunit, respectively. The plasmid contains kanamycin resistance gene for selection in bacteria. The plasmid backbone is optimized for efficient growth in bacteria.
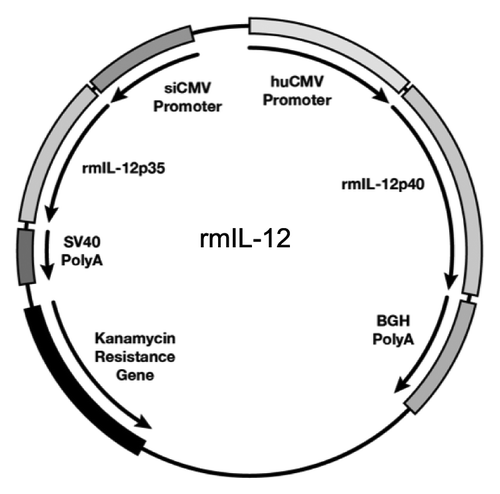
Figure 2. In vivo bioactivity of optimized rmIL-12 DNA injected in rhesus macaques via intramuscular injection. (A, B) Plots from individual rhesus macaques depicting overlays of plasma macaque IL-12p40 (black solid line; left Y-axis) and IFN-γ (dotted line; right Y-axis). Macaques (n = 4) were injected intramuscularly followed by in vivo electroporation with either 0.1 mg (A, left upper panels) or 0.4 mg (B, right upper panels) of optimized rmIL-12 DNA or sham DNA (lower right and left panels; mean and SEM are shown). The animals in panel A were injected 3 times with intervals of 2 mo, respectively. The animals in panel B were injected twice with an interval of 9 weeks. Arrows mark the day of the injection. (C, D) The plots show the difference between basal levels (day 0; prior to injection) and at day 5 of plasma IL-12p40 (upper panels) and the peak levels of plasma rmIFN-γ (lower panels) after injection of 0.1 mg (C, left panels) or 0.4 mg (D, right panels) rmIL-12 DNA. Note for the 0.1 mg IL-12 DNA group at EP1 (left upper panel), the day 4 measurements were used since the day 5 samples were not collected. For rmIFN-γ, the peak values are shown. The basal levels of rmIFN-γ were below the threshold of detection in all macaques. The mean values are shown.
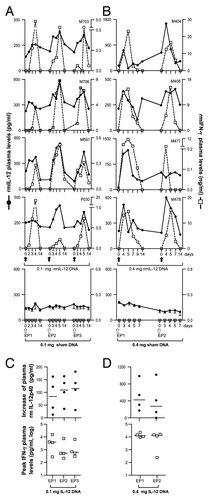
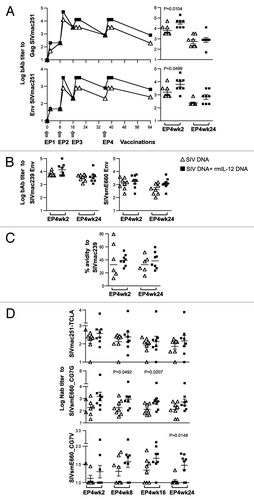
Figure 3. The IL-12 DNA-adjuvanted vaccine group developed higher cytotoxic T cell responses. (A) The outline of the SIV vaccine study is shown. Two groups of macaques (n = 8 each) were subjected to intramuscular injection followed by in vivo EP using a mixture of SIV DNAs in the absence or presence of 0.1 mg of the optimized macaque IL-12 DNA plasmid (AG157) at 0, 8, 16, and 36 weeks. (B) Cellular immune responses to SIV Gag and SIV Env were measured from individual animals at 2 weeks post EP2 and 2 and 24 weeks post EP4 using polychromatic flow cytometry. Total (Env + Gag) SIV-specific IFN-γ+ T cells (upper panel) and the corresponding Granzyme B+ (GzmB+) cytotoxic T cell subpopulation (middle panel) and GzmB- T cell subpopulation (lower panel) are shown for the two groups of macaques. The bars represent the mean values and standard error of mean (SEM). Mann-Whitney non-parametric t-test was employed to compare the 2 groups using Prism software (GraphPad Software, Inc.). The significant p values are shown.
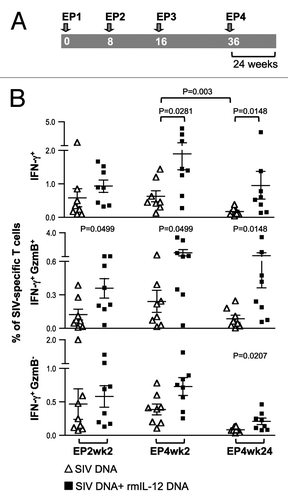
Figure 4. Comparison of the Gag- and Env-specific CD4+, CD8+ and CD4+CD8+ double-positive (DP) T cell subsets. (A, B) Gag (A)- and Env (B)-specific IFN-γ + producing CD4+, CD8+ and CD4+CD8+ DP T cell populations were analyzed from individual animals by flow cytometry at EP2wk2, EP4wk2 and EP4wk24. The data are shown as % of total parent CD4+, CD8+ or CD4+CD8+ T cells. The bars represent mean values and SEM.
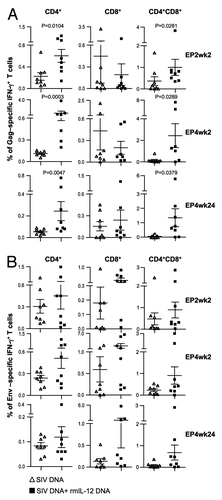
Figure 5. Comparison of the Gag- and Env-specific memory T cell subsets. (A, B) The frequency of (A) Gag-specific GzmB+ CD4+ and CD4+CD8+ DP transitional memory (TM) and effector memory (EM) T cells and (B) Env-specific GzmB+ CD8+ and CD4+CD8+ DP TM and EM T cells as analyzed by flow cytometry from individual animals at EP2wk2, EP4wk2 and EP4wk24 are shown. The SIV-specific IFN-γ producing GzmB+ cells are shown as % of total Gag- or Env-specific T cells. The mean and SEM values are shown. The significant p values are shown.
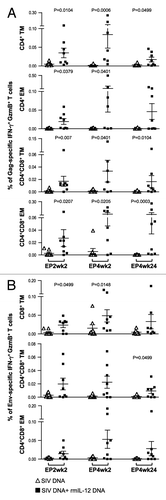
Figure 6. The IL-12 DNA-adjuvanted group developed broader humoral immune responses. (A) Endpoint titers of binding antibodies to SIVmac251 Gag (upper panel) and SIVmac251 Env (lower panel) were measured in pooled plasma samples (left panels). The right panels show the data from individual animals at 2 and 24 weeks post EP4. The mean and SEM values are shown. (B) Endpoint titers of binding antibodies to SIVmac239 (left panels) and SIVsmE660 (right panels) Env were measured in individual animal at 2 and 24 weeks post EP4. (C) Avidity index of the SIVmac239 Env bAb. The avidity of two of the 8 animals in the un-adjuvanted group could not be assessed at both time points. The antibody binding titers and avidity index were determined from parallel plates. (D) Neutralizing antibody titers from individual plasma samples from the vaccinated macaques against SIVmac251-TCLA (upper panel), and against the SIVsmE660 BR-CG7G (middle panel) and BR-CG7V (lower panel) Env proteins at 2, 8, 16 and 24 weeks post EP4 are shown. The bars represent mean and SEM values.