Figures & data
Figure 1. Unrooted Pol sequence neighbor joining (NJ) dendrogram of the retroviral genera: α-, β-, gamma-, delta-, epsilon-, lenti- and spuma retroviruses.Citation76 The viruses used in this comparative study are indicated.
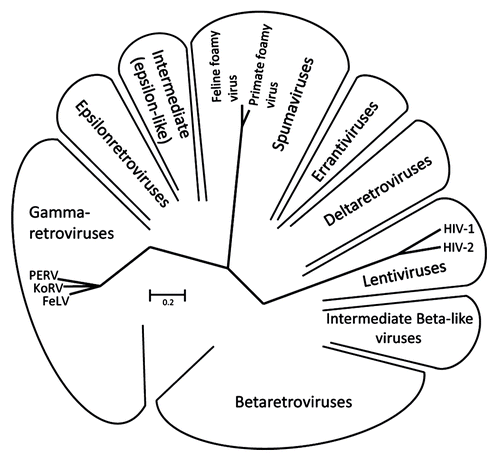
Figure 2. Main functional domains of the retroviral TM proteins (NHR - N-terminal helical region, C - C - Cys-Cys-loop, CHR - C-terminal helical region). The TM proteins of PERV, FeLV, HIV and FFV are presented. C indicates a cysteine, a star indicates a potential N-linked glycosylation site.
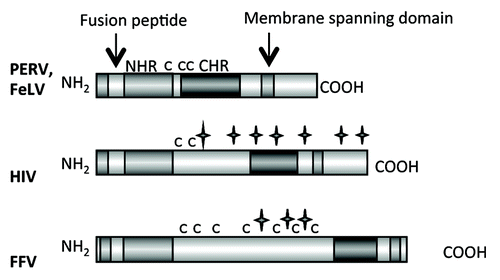
Table 1. The transmembrane envelope proteins of different retroviruses
Figure 3. Flowchart of the expression and purification process for the generation of TM proteins for immunisation and testing the induced sera for binding and neutralizing antibodies.
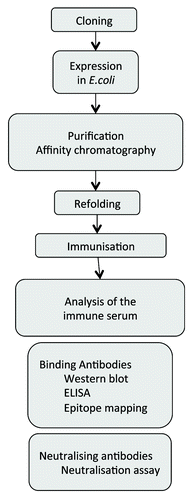
Figure 4. (A) Schematic presentation of the ectodomain of p15E and (B) localization of the epitopes recognized by sera induced by immunisation with the ectodomain of the TM protein of three gammaretroviruses, PERV, FeLV and KoRV in different species. FPPR — fusion peptide proximal region, MPER — membrane proximal external region. Epitopes strongly recognized by sera from all immunised animals are in dark gray, epitopes recognized less and only by sera from some immunised animals are in light gray. The number of sera analyzed by epitope mapping are indicated.
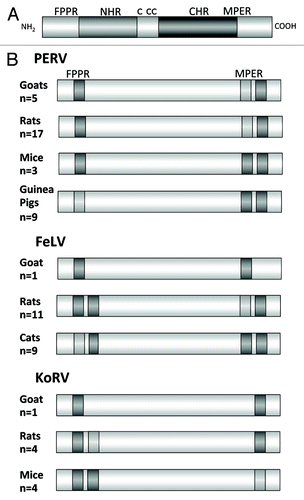
Figure 5. Location of the epitopes recognized by immune sera from rats immunised with the recombinant ectodomain of gp41 of HIV-1 and gp36 of HIV-2. Epitopes strongly recognized by sera from all immunised animals are in dark gray, epitopes recognized less and only by sera from some immunised animals are in light gray.

Figure 6. Generation of hybrid proteins consisting of a backbone derived from the TM protein p15E of PERV and the FPPR and MPER of gp41 of HIV-1, which replace the FPPR and MPER of p15E. The FPPR and MPER of p15E were removed and the FPPR and MPER (containing the epitopes of the broadly neutralizing antibodies 2F5 and 4E10) of gp41 of HIV-1 were inserted. NHR - N-terminal helical region, C - C or C - CC - Cys-Cys-loop, CHR - C-terminal helical region, MSD - membrane spanning domain.
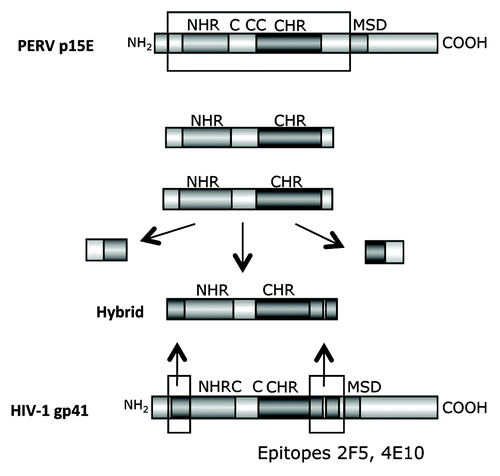
Figure 7. (A) Structure of the hybrid protein used for immunisation. The hybrids were generated as described in . For abbreviation see and . The origin of the fusion peptide (FP) and the membrane spanning domain (MSD), either from gp41 of HIV-1 or from p15E of PERV are indicated. Hybrid 3 and 4 lack the FPPR and MSD, in hybrid 4 the FPPR region was shifted. (B) Localization of the epitopes recognized by sera from animals immunised with the different hybrid proteins. The first part of the sequence corresponds to the FP/FPPR sequence, the second part to the MPER. The sequences of the epitopes of 2F5 and 4E10 are in bold, the sequences of the peptides shown to enhance the binding of 2F5 and 4E10 to their epitopes when interacting are underlined.
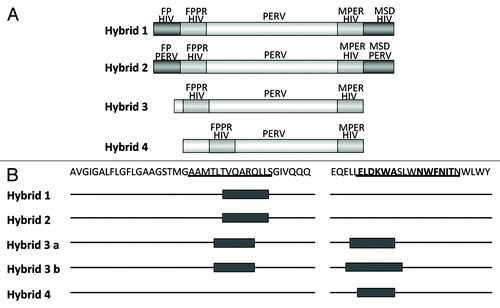
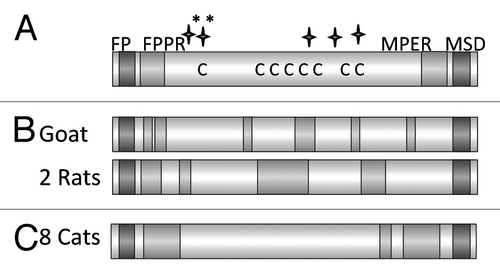
Figure 8. (A) Schematic presentation of the main domains of the TM protein of the feline foamy virus (FFV). Abbreviations see . C marks the cysteine residues, long stars the N glycosylation sites, long stars marked with * the potential N glycosylation sites. (B) Localization of the epitopes recognized by the immune sera induced by immunisation with the TM protein of FFV. Sera from one goat, and two rats which were immunised with the TM protein produced in bacteria were analyzed. (C) Localization of the epitopes recognized by the sera from 8 cats naturally infected by FFV.