Figures & data
Figure 1. The rapid emergence of H7N9 in several locations in China reflects wide-spread distribution in the animal population and potential for even greater spread. H7N9 is a virus that deserves serious consideration. Should a traveler get infected in China, flight routes from the outbreak regions would quickly carry any human-transmissible virus to huge population centers in Europe, North America and Asia. An estimated 70% of the world population resides within two hours’ travel time of destination airports (calculated using gridded population-density maps and a data set of global travel times, map supplied by A. J. Tatem, Z. Huang and S. I. Hay (2013). Unpublished data. (A.J.T., University of Southampton, UK; Z.H., University of Florida, Gainesville; S.I.H., University of Oxford, UK.) Reprinted by permission from Macmillan Publishers Ltd: Nature News 2013.Citation9
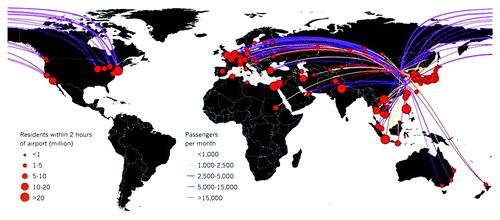
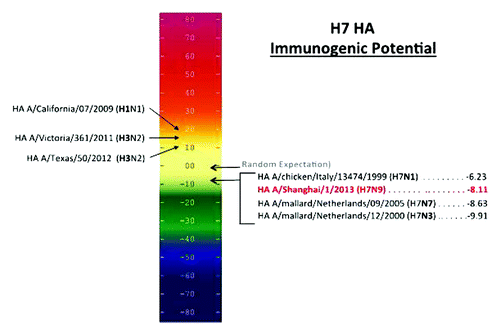
Figure 2. Potential immunogenicity of emerging influenza A (H7N9) HA and HA from the current seasonal vaccine and H7 vaccines. The number of HLA ligands (putative T cell epitopes) per unit protein is plotted on an Immunogenicity Scale. This scale is correlated with observed immunogenicity in retrospective and prospective studies.Citation16,Citation17 The numbers used in this scale reflect the difference between the number of predicted T cell epitopes we would expect to find in a protein of any given size by chance alone (based on an evaluation of more than 10,000 random protein sequences) and the number of putative epitopes predicted by the EpiMatrix System for a given protein. The EpiMatrix Protein Score of an “average” protein is zero. EpiMatrix Protein Scores above zero indicate the presence of excess MHC ligands and denote a higher potential for immunogenicity, while scores below zero indicate the presence of fewer potential HLA ligands than expected and a lower potential for immunogenicity. H7 has fewer HLA ligands than expected, which is reflected in its negative immunogenicity score. Seasonal strains of influenza are scored for potential immunogenicity on the left; their scores are much higher than H7N9.