Figures & data
Figure 1. Informatic-driven identification of influenza H1N1 hemagglutinin and neuraminidase immunogenic consensus sequences. Results of the step-wise computational process of screening H1-HA and N1-NA sequences for conserved and potentially immunogenic epitopes and constructing immunogenic consensus sequences are shown.
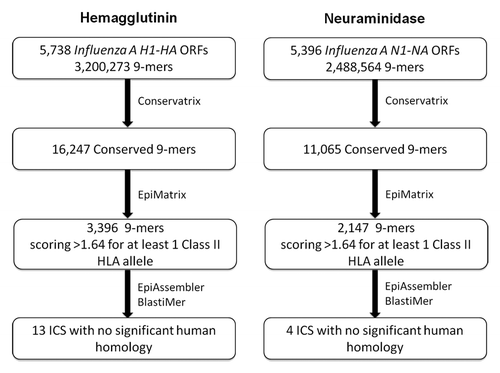
Figure 2. Conservation distribution of influenza H1N1 hemagglutinin and neuraminidase 9-mers. Pre-pandemic and pandemic H1-HA (A) and N1-NA (B) sequences were parsed into overlapping nine amino acid frames with a one amino acid frameshift, and unique 9-mers were evaluated for percent coverage of source antigens at 100% sequence identity using the Conservatrix algorithm. Inset: Close-up of the 75–100% coverage range.
Figure 3. Immunogenic consensus sequence influenza H1N1 strain coverage and HLA binding potential. ICS construction yielded 13 H1-HA and four N1-NA sequences with > 85% coverage of input proteins and EpiMatrix cluster scores > 10, representing significant HLA binding potential. Bars represent EpiMatrix cluster score and open circles percent strain coverage.
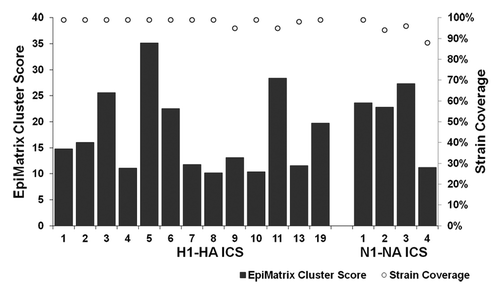
Table 1. H1N1 immunogenic consensus sequences
Figure 4. ICS peptide – HLA DR binding affinities. Peptide identifiers and sequences are noted in the first and second columns, respectively. IC50 values in μM units were calculated from curves fitted to dose-dependence competition binding data for each peptide-HLA DR allele pair. Peptide binding affinity is shown according to the following classification: IC50 < 0.1 µM (black), 0.1 µM < IC50 < 1 µM (dark gray), 1 µM < IC50 < 10 µM (gray), 10 µM < IC50 < 100 µM (light gray), IC50 > 100 µM (lightest gray). IC50 values too high to accurately measure under binding conditions tested are considered non-binders (NB; shown in white cells).
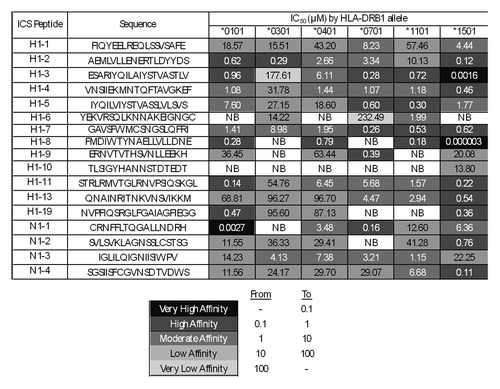
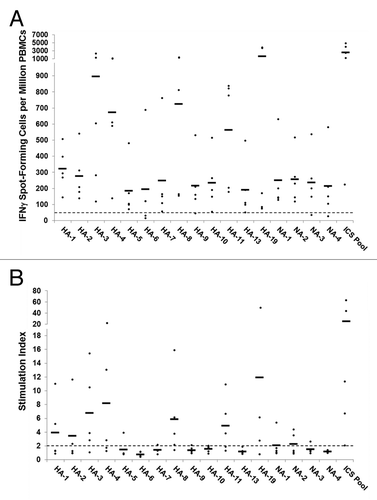
Figure 5. Antigen-specific human IFNγ ELISpot responses to computationally identified influenza HA and NA immunogenic consensus sequences. ICS were assayed for T cell reactivity by IFNγ ELISpot assay using PBMCs isolated from normal human donors (n = 5). An ELISpot response was considered positive if three criteria were met: (1) spot-forming cells (SFC) per million PBMC were at least 50 over background; (2) SFC per million PBMC were at least 2-fold over background; and (3) antigen-stimulated SFC numbers were statistically different (p < 0.05) from non-stimulated counts. (A) The numbers of SFC over background per million PBMCs that secrete IFNγ in response to individual and pooled influenza HA and NA ICS are presented. Individual subject responses are represented by dots and the average response across subjects by horizontal lines. The 50 SFC over background per million PBMCs cutoff is denoted by the dotted line. (B) The ELISpot response stimulation index, representing the ratio of antigen-stimulated SFC counts to non-stimulated counts, is presented. Stimulation index values per individual subject are represented by dots and the average values across subjects by horizontal lines.