Figures & data
Table 1. Results of polymorphisms association analysis in 18 HIV+ patients underwent DC-based immune treatment against HIV-1 classified in good responder (GR) and weak or transient responder (WTR) according to Lu et al.Citation3
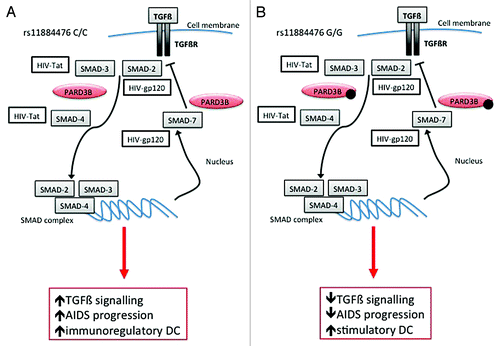
Figure 1. Plasma viral load (PVL) reduction and cellular response in 18 HIV+ patients who’s underwent DC-based immune treatment against HIV-1 according to PARD3B rs11884476 genotypes. Change in PVL expressed as log change (∆log), change in CD4+ and CD8+ cells counts (∆cells/µl) and change in percentage of CD4+ and CD8+ cells producing IFN-ϒ (∆%) are reported for the 18 HIV+ patients included in the phase I clinical trial of DC-based immune-therapyCitation3 classified according to PARD3B rs11884476 genotypes. The data, obtained from Lu et al.,Citation3 represent difference (∆) between values presented 1 y after immunization and before the starting of the trial. Individual data and media were reported. (A) Plasma viral load (PVL). Individual blood CD4+ (B) and CD8+ (C) cell counts. (D and E) Intracellular IFN-γ detection of T cells following stimulation with HIV-1-pulsed DC. Percentage of total CD4+ (D) or CD8+ (E) cell secreting IFN-γ is reported. T test analysis was performed between C/C and C/G groups and between C/C+G/G and C/G groups according to an over-dominant model. Being unique value the G/G has been excluded from the analysis. *P < 0.05; **P < 0.01.
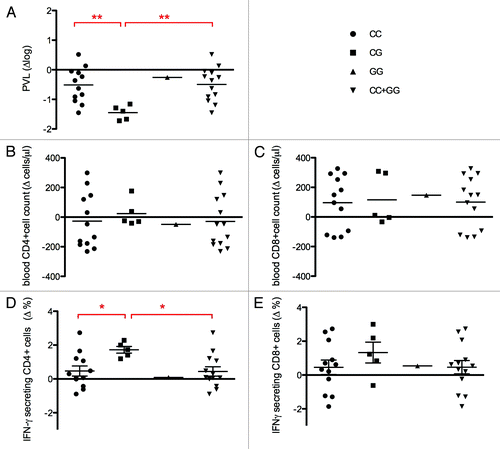
Figure 2. Possible interactions between PARD3B and SMAD proteins influencing TGFß signaling. The two hypotheses concerning the dual role of PARD3 in terms of viral replication control or DC-mediated lymphocytes activation are reported according to rs11884476 genotypes (2A: wild type C/C genotype; 2B: G/G genotype). The up or downregulation of TGFß signaling and the consequences in terms of AIDS progression or DC immune-regulation are evidenced in red rectangles.