Figures & data
Table 1. Sample information and clinical symptoms of flu infection among A(H1N1)pdm09 Taiwan patients
Figure 1. New genetic variants among A(H1N1)pdm09 isolates collected at Taiwan between 2009–2011 detected by MSSCP genotyping. RT-PCR products of hemagglutinin gene obtained from pandemic Taiwan A(H1N1)pdm09 virus isolates, as well as reference seasonal (s) and pandemic (p) strains of influenza virus A(H1N1)pdm09 were denatured and ssDNA’s were separated on a 9% polyacrylamide gel using MSSCP method under optimum electrophoretic conditions. DNA bands were visualized with silver stain. Strains are indicated as follows: s – reference seasonal strain, p – reference pandemic strains. Taiwan isolates are described with symbols listed in . Note the presence of five distinct MSSCP electrophoretic profiles (arrows at the bottom of the figure) (samples number: 2010–03994, 2011–01219, 2010–01164, 2010–05270, 2010–05347) among samples, which based on RT-PCR assay, were classified as A(H1N1)pdm09 strain. Dividing lines indicate grouping of images from different parts of the same gel.
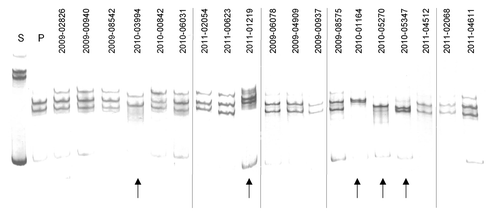
Table 2. Genetic diversity of HA gene fragment in Taiwan A(H1N1)pdm09 isolates
Figure 2. Schematic representation of genetic diversity of hemagglutinin (HA) sequence in five Taiwan isolates of A(H1N1)v pan09 strain. Black arrows above and below A(H1N1)v pdm09 reference sequence indicate modified DNA codons. Red letters show nucleotide changes. Altered amino acids in HA protein sequence are also shown. Blue star over the description indicates amino acids localized in the epitope E of HA. Additionally, red arrows underline three point mutations, which are translated to amino acids in the epitope E of HA.
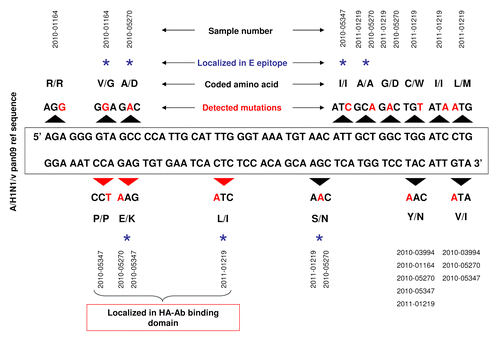
Figure 3. Structure of HA-Ab complex. Hemagglutinin protein monomer (blue color) interacting which neutralizing antibody (red color). Amino acid 66, localized on the HA-Ab interface is marked in green.
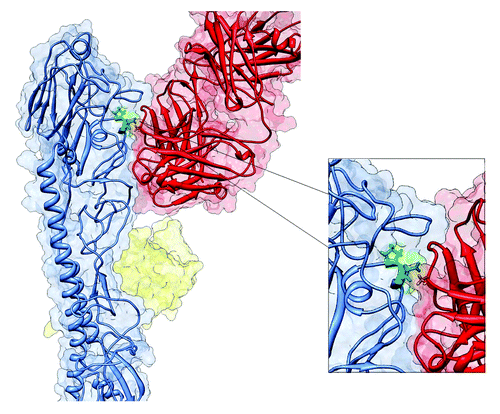
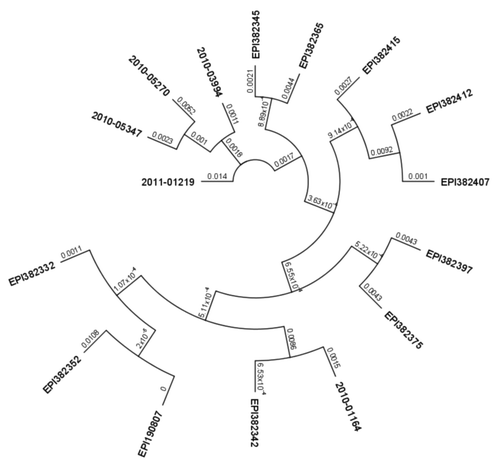
Figure 4. Phylogenetic tree of nucleotide sequences of hemagglutinin gene of Taiwan isolates of influenza A(H1N1)pdm09 virus. All sequences included in phylogenetic analysis were 935 nucleotides long. All Taiwan isolates deposited from 2009 through 2011 were retrieved from the EpiFlu Database. Each isolate (EPI210250 HA, EPI382332 HA, EPI382342 HA, EPI382415 HA, EPI190807 HA, EPI382397 HA, EPI382345 HA, EPI382352 HA, EPI382407 HA, EPI382412 HA, EPI382365 HA, EPI382375 HA) represents a number of identical sequences used in alignment. Isolates studied in this paper are described in . Numbers on branches indicate number of nucleotide substitutions per site.