Figures & data
Figure 1. Phylogenetic relationship of meningococcal serogroup B fHBP variants. Figure adapted from Murphy et al.Citation20 Phylogenetic tree of fHBP/LP2086 variants based on clustalW alignment and drawn with MEGA 4. Highlighted are subfamily A and B, the subgroups within the respective subfamilies, and the components of the bivalent rLP2086 investigational vaccine, fHBP variants A05 and B01. During the development of bivalent rLP2086, alternative nomenclature systems were used for fHBP in the scientific literature.Citation21,Citation38 The scale bar indicates phylogenetic distance based on protein sequence. The phylogenetic distance between any two variants is the length of the line traced from one branch back to the first common node on the tree and along the branch to the second variant. The end of each branch on the tree represents a specific fHBP variant. The longer the line connecting the two variants, the greater is the amino acid sequence diversity between them.
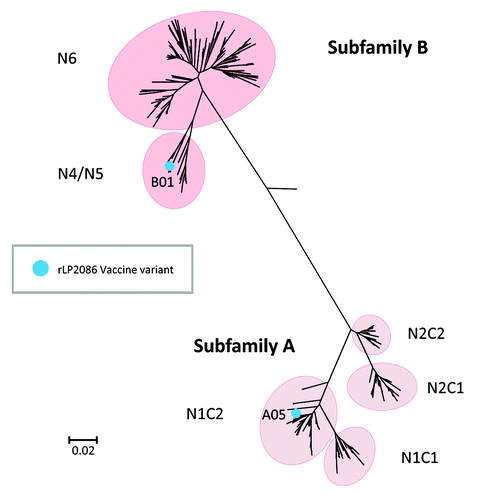
Table 1. Immunization of mice with rLP2086-B01 elicits greater bactericidal activity than rP2086-B01 using a MnB SBA test strain that expresses heterologous (to vaccine antigen) fHBP variant B24
Table 2. Immunization of rhesus macaques with rLP2086-A05 elicits greater hSBA responses than rP2086-A05 using MnB strains that express homologous or heterologous (to vaccine antigen) subfamily B fHBP variants
Table 3. The composition and fHBP subfamily distribution of the MnB SBA strain pool (2000–2006)
Figure 2. Distribution of fHBP subfamily A or b expressing strains stratified by patient age in the MnB SBA strain pool. Figure adapted from Hoiseth et al.Citation22 The distribution of fHBP subfamily A and subfamily B expressing strains within different age groups in the MnB SBA strain pool (age data available for 1215 subjects). Age groups with significant differences (*P < 0.0005) and with highly significant differences (**P < 0.0001) in subfamily distribution compared with the <1 y age group are marked (Fisher Exact Test).
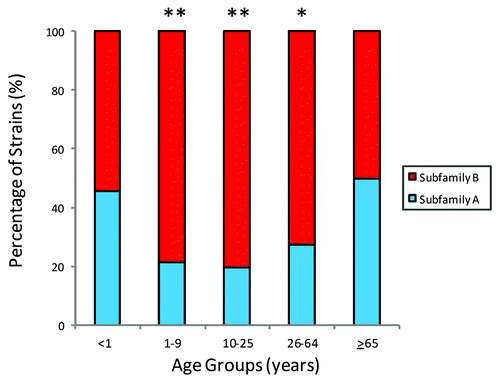
Figure 3. Prevalence of fHBP subfamily A and B variants in the MnB SBA strain pool and the US subset of the MnB SBA strain pool. Data taken from Murphy et al.Citation20 The ten most prevalent fHBP variants (each with ≥20 representative strains) and the cumulative percentage of strains in the MnB SBA strain pool (n = 1263) are pictured. The prevalence of these ten fHBP variants in the US subset of the MnB SBA strain pool (n = 432) and the non-US subset of the MnB SBA strain pool (n = 831) are shown.
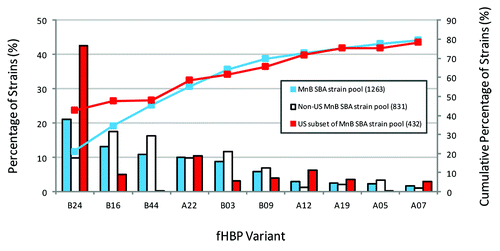
Figure 4. The fHBP variant distribution within the major clonal complexes of the MnB SBA strain pool. Data taken from Murphy et al.Citation20 The relative abundance and fHBP variant distribution within the ten most prevalent CCs in the MnB SBA Strain Pool (n = 1263) is shown. Each color in the respective columns corresponds to an individual fHBP variant and the prominent variants are labeled. Above each of the columns is the number of different fHBP variants that were found associated with the respective CCs.
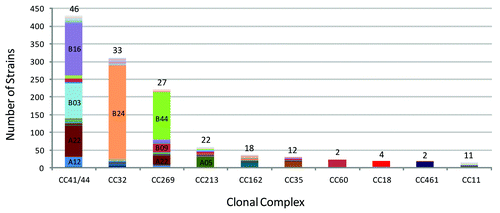
Figure 5. Overview of the MEASURE assay. Bacteria are grown following standard hSBA procedures. Briefly, MnB bacteria are streaked onto plates and grown overnight before inoculating liquid media with colonies and growing to a specific OD. Bacteria are then stained using a 3-step antibody staining method. Bacteria were stained with the primary anti-LP2086 antibody, MN86–994–11–1, followed by a biotinylated mIgG secondary antibody and tertiary PE-Streptavidin to amplify the fluorescence signal and increase the dynamic range.
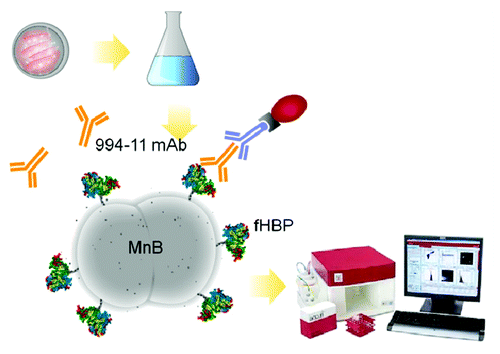
Figure 6. fHBP surface expression correlates with strain susceptibility in the SBA. Data adapted from Jiang et al.Citation19 Median fluorescence intensity (MFI) was determined using the validated MEASURE assay (McNeil et al., manuscript in preparation). Invasive MnB strains (n = 100) were tested using the Measure assay and the MFI was plotted. Each strain was also run in the hSBA assay. Red squares represent MnB strains that were not killed in the hSBA and blue triangles represent MnB strains that were killed.
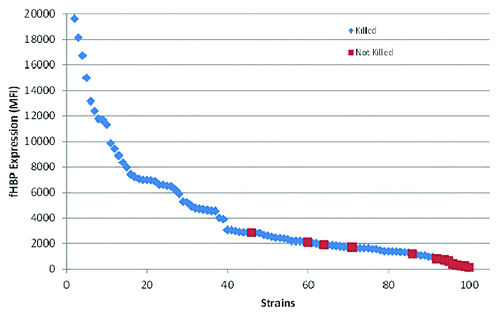
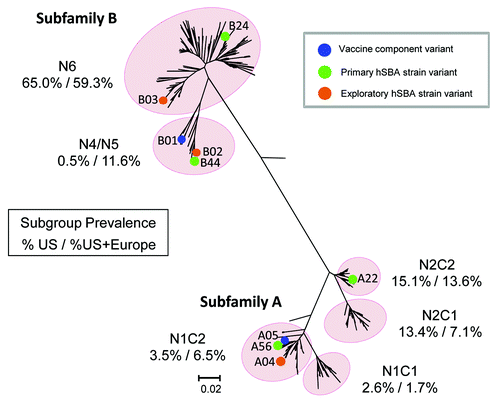
Figure 7. fHBP Phlyogenetic Tree Illustrating MnB hSBA Strain and Vaccine Component Variants. Phylogenetic tree of fHBP/LP2086 variants in the MnB SBA strain pool based on clustalW alignment, and drawn with MEGA 4. Highlighted are the fHBP variant components of bivalent rLP2086 (A05 and B01), the fHBP variants expressed by the primary hSBA strains, and the fHBP variants expressed by the exploratory hSBA strains listed in . The numbers beneath each fHBP subgroup in the figure represent the percentage of isolates in the US subset of the MnB SBA strain pool (on the left) or the MnB SBA strain pool (on the right) that reside in each subgroup. The scale bar indicates phylogenetic distance based on protein sequence.