Figures & data
Figure 1. STAT3 with DNA-consensus sequence. STAT3 monomer showing the N-terminal coiled-coil domain, the DBD (half site), the SH2 domain and the C-terminal domain. Basic residues are blue and acid ones are red. The STAT3 crystal coordinates were downloaded from the protein data bank (PDB, file: 1BG1) and analyzed using the chimera program.Citation73 Note that the model shown comprises residues 136 to 716;Citation74 hence, the protein’s N-terminal and C-terminal domains (comprising the transactivation domain) are missing; the coordinates corresponding to the cDNA strand were missing in the model and had to be reconstructed.Citation65
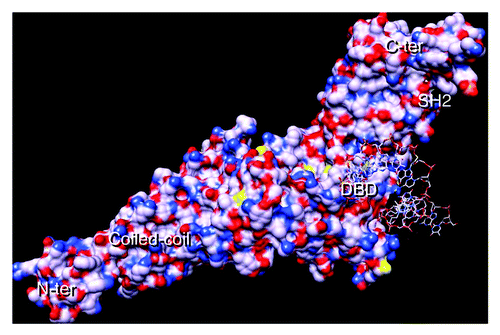
Figure 2. STAT3 activation. The transcription factor STAT3 is present in a latent inactive non-phosphorylated form in the cytoplasm. Activated cytokine receptors activate the kinases JAK, which phosphorylate tyrosines located in the cytoplasmic portion of the cytokine receptors creating STAT-binding motifs. Once bound to these motifs, STAT3 becomes in turn phosphorylated by the JAKs. The phospho STAT3 dimers (colored in pink) enter the nuclei and bind STAT3 target genes; note that the DNA-bound dimer is drawn differently to indicate the STAT3 conformational change suggested by molecular dynamics simulations.Citation22 STAT3 can also be activated by tyrosine kinases of the Src family (SFK), the SFKs can themselves be activated by G protein-coupled receptors (GPCR) or growth factor receptors, the growth factor receptors (including EGF-receptors and VEGF-receptors) can also phosphorylate STAT3. The DBD is created by the phosphotyrosine 705/SH2-mediated dimer. There are also monomers, and dimers with non-phosphorylated STAT3, which can shuttle between the cytoplasm and the nucleus.
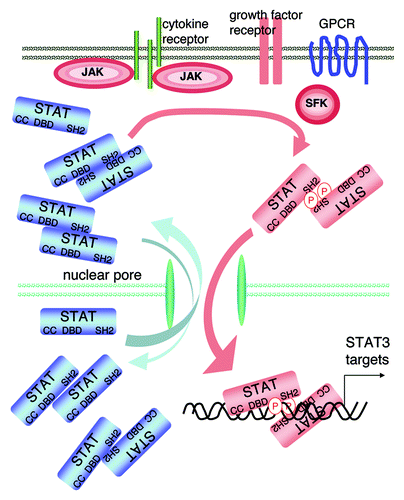
Figure 3. STAT3 nuclear entry. The tyrosine phosphorylated STAT3 dimer interacts with the importins through its two NLSs [one within the coiled-coil (mL) domain: arginines 214 and 215; one within the DBD: arginines 414 and 417]. This complex is carried through the nuclear pore complex (NPC) by the Ran GDP which is formed in the cytoplasm by hydrolysis of Ran-bound GTP by GTPase activating protein (GAP). The NPC comprises nucleoporins (Nup). The importins release the dimer once it enters the nucleus. The high level of Ran-GTP in the nucleus is the result of a high GTP-exchange factor (GEF) activity in this compartment. The STAT3 dimer interacts with the STAT3 DNA consensus motif and then is released; once released from DNA the dimer is dephosphorylated by a nuclear phosphatase (PtPase). The dephosphorylated STAT3 is exported to the cytoplasm in combination with exportins and Ran-GTP.
![Figure 3. STAT3 nuclear entry. The tyrosine phosphorylated STAT3 dimer interacts with the importins through its two NLSs [one within the coiled-coil (mL) domain: arginines 214 and 215; one within the DBD: arginines 414 and 417]. This complex is carried through the nuclear pore complex (NPC) by the Ran GDP which is formed in the cytoplasm by hydrolysis of Ran-bound GTP by GTPase activating protein (GAP). The NPC comprises nucleoporins (Nup). The importins release the dimer once it enters the nucleus. The high level of Ran-GTP in the nucleus is the result of a high GTP-exchange factor (GEF) activity in this compartment. The STAT3 dimer interacts with the STAT3 DNA consensus motif and then is released; once released from DNA the dimer is dephosphorylated by a nuclear phosphatase (PtPase). The dephosphorylated STAT3 is exported to the cytoplasm in combination with exportins and Ran-GTP.](/cms/asset/6e103a71-4d84-413c-85b0-e0f6cbd1f78d/kjks_a_10922882_f0003.gif)
Table 1. Comparison and alignment of the STAT3 and STAT1 specific DNA recognition sites and of the dODNs used to inhibit STAT3
Figure 4. A detailed view of STAT3 SH2 and DBD regions. (A) STAT3 surface obtained as in . The areas in squares are the SH2 region where small molecules interact (B and C) and the SH2/DBD region where G quartets interact (D and E). (B) Close up view of the region of the SH2 of STAT3 that interacts with the small molecule inhibitors: the key aminoacids involved in interaction are labeled; the inhibitors interact particularly in the groove located between arginine 595 and lysine 591. (C) The same area as in (B) is shown but STAT3 and STAT1 are superimposed; this shows the overall great similarity between these two regions, yet some differences are present, accounting for the capacity of the inhibitors to discriminate between STAT3 and STAT1 (arrows); STAT1 crystal coordinatesCitation66 used in (C) were from PDB file 1BF5. (D) Modeled interaction of inhibitor S31-201 with STAT3 SH2 domain, note the important role of lysine 591, serine 611 and arginine 609 in the interaction. (E) Same as (D), with added phosphotyrosine-peptide, showing its overlap with the STAT3 SH2/small molecule inhibitors binding area. (F) Detailed view of the region of STAT3 interacting with G quartets, this region includes the phosphotyrosine 705-interacting region of SH2 and a neighboring region including part of the DBD, including glutamic acid 638, glutamine 644, aspartic acid 647, glutamine 643 and asparagine 646. (G) Same region as in F is shown with the superimposition of STAT1, the major differences are indicated by arrows. [Panels (D and E) are reprinted from ref. Citation44 with permission; © 2007 National Academy of Sciences USA.]
![Figure 4. A detailed view of STAT3 SH2 and DBD regions. (A) STAT3 surface obtained as in Figure 1. The areas in squares are the SH2 region where small molecules interact (B and C) and the SH2/DBD region where G quartets interact (D and E). (B) Close up view of the region of the SH2 of STAT3 that interacts with the small molecule inhibitors: the key aminoacids involved in interaction are labeled; the inhibitors interact particularly in the groove located between arginine 595 and lysine 591. (C) The same area as in (B) is shown but STAT3 and STAT1 are superimposed; this shows the overall great similarity between these two regions, yet some differences are present, accounting for the capacity of the inhibitors to discriminate between STAT3 and STAT1 (arrows); STAT1 crystal coordinatesCitation66 used in (C) were from PDB file 1BF5. (D) Modeled interaction of inhibitor S31-201 with STAT3 SH2 domain, note the important role of lysine 591, serine 611 and arginine 609 in the interaction. (E) Same as (D), with added phosphotyrosine-peptide, showing its overlap with the STAT3 SH2/small molecule inhibitors binding area. (F) Detailed view of the region of STAT3 interacting with G quartets, this region includes the phosphotyrosine 705-interacting region of SH2 and a neighboring region including part of the DBD, including glutamic acid 638, glutamine 644, aspartic acid 647, glutamine 643 and asparagine 646. (G) Same region as in F is shown with the superimposition of STAT1, the major differences are indicated by arrows. [Panels (D and E) are reprinted from ref. Citation44 with permission; © 2007 National Academy of Sciences USA.]](/cms/asset/8396b907-aa89-431b-bff9-a8d14573cff6/kjks_a_10922882_f0004.gif)
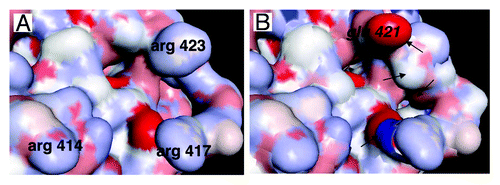
Figure 5. Detailed view of the area of STAT3 interacting with DNA and importins. (A) Detail of the area of STAT3 interacting with the DNA consensus sequence. Arginines 414 and 417 play a key role in the interaction of STAT3 with importins and nuclear translocation, arginine 423 is involved in the interaction of DNA with STAT3.Citation65 (B) The same area as in (A) is shown, the corresponding area of STAT1 has been superimposed, showing the high level of homology between STAT3 and STAT1, except for glutamic acid 421 (in italic) of STAT1 whose interaction with DNA is very different from that of arginine 423 of STAT3 (see ref. Citation65), and other differences indicated by arrows.