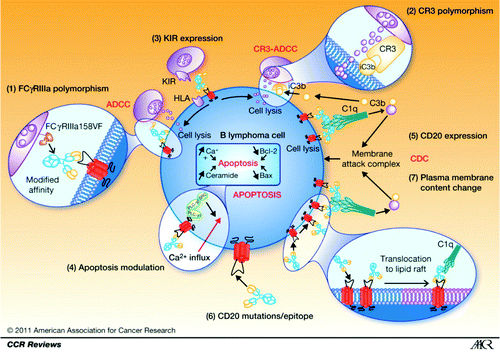
Figures & data
Figure 1. Mechanism of rituximab action, resistance, and causes of variability of response. (1) FCGR3A-158FV genotype affects the affinity of rituximab for human IgG1. (2) Rituximab binds more strongly to homozygous FcγRIIIa-158V effector cells than to FcγRIIIa-158F carrier effector cells, resulting in significantly different response rates to rituximab. (3) Polymorphism located in the C3-interacting site of CR3/CD11b has been shown to be associated with PFS in patients with follicular lymphoma treated with rituximab alone. (4) NK cell target recognition depends mainly on the surveillance of HLA class I molecules by B-cells interacting with KIRs on NK cells modulating ADCC and CR3-ADCC. (5) Apoptosis as an effector mechanism of rituximab has been shown to be modulated, resulting in in vitro resistance to both rituximab and chemotherapy. (6) CD20 expression influences CDC induced by rituximab. (7) A rare cause of rituximab resistance may be mutations of the CD20 gene, resulting in reduced CD20 expression or affinity for rituximab. Antitumor effects of rituximab are dependent on translocation of antibody-bound receptor to lipid rafts in the plasma membrane. All changes in the cholesterol content of the plasma membrane can thus affect the lipid-raft–dependent activity of rituximab. Reprinted with permission from Cartron G et al. Clin Cancer Res 2011; 17:19–30.