Figures & data
Figure 1. Fc structure (A) indicating the locations of structural domains, two attached oligosaccharides colored cyan, and the position of Gly237 residues used to calculate the CH2-CH2 distance. For chain H, the CH2 domain is red and the CH3 domain is orange. For chain K, the CH2 domain is green and the CH3 domain is blue. The hinge, which was present in all glycoform variant simulations, has been left out of this representation for the sake of clarity. CH2 domain schematic (B) with β-strands labeled and residue indices for strands in Kabat numbering scheme. Two phenylalanine residues 241 and 243, important for CH2 domain stability, are located on β-strand A. The oligosaccharide attachment residue, Asn297, is located in loop C’E.
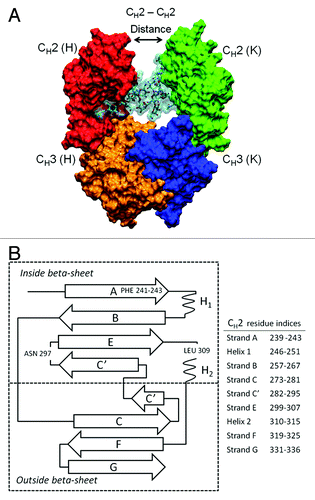
Figure 2. Structure, composition, and branches are labeled for oligosaccharides of G2FS2. The glycan has three terminal carbohydrates: FUC-2, and two N-acetyl neuraminic acids or sialylic acids, labeled S. Desialylating G2FS2 produces G2F. Alternate glycan compositions and structures are shown using a simplified notation. Note that MN2, not shown, can be formed by removing MAN5 and MAN8 from M3N2.
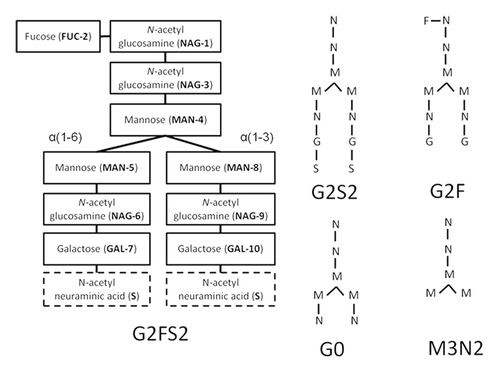
Table 1. Simulations performed in this study
Table 2. Ensemble averages, standard deviations, and standard error of the mean
Figure 3. The average CH2-CH2 distance during simulations at (A) 300 K and (B) 375 K is plotted. Distances are averaged over a 5 ns window and plotted in the window center. Even though glycoform variant starting structures are the same, initial CH2-CH2 distances for the three glycoform variants are slightly different because the first data points reported are at 2.5 ns due to averaging. Figure insets (α) and (β) represent DEGLYCO structures along the trajectory. At 300 K, simulations began with DEGLYCO in an open conformation. As DEGLYCO simulations progressed, the CH2-CH2 distance decreased and the FcR binding site closed. At 375 K, the CH2-CH2 distance increased as CH2 domains collapsed onto CH3 domains. The FcR binding site was also closed in the collapsed structure.
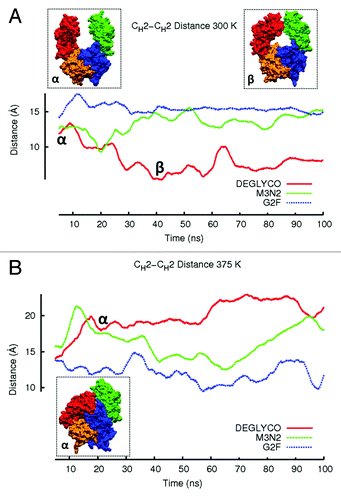
Figure 4. Residue-wise RMSDs for all residues of CH2 domains calculated in simulations at (A) 300 K and (B) 375 K. Residue-wise RMSD values for each glycoform variant were averaged between the two independent simulation runs at each temperature and then averaged over a 5 ns window. Local structure including β-strands and loops are illustrated with black bars.
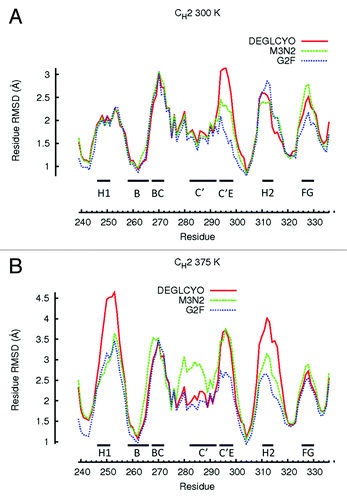
Table 3. Glycan contacts for G2F and M3N2 at 300 K
Figure 5. RMSF values are plotted for individual carbohydrates of G2F and M3N2. RMSF values were averaged between the two independent simulation runs at each temperature. RMSF values for data points simulated at 300 K are connected by lines. RMSF values for simulations preformed at 375 K are not connected by lines due to the absence of terminal GAL and NAG carbohydrates.
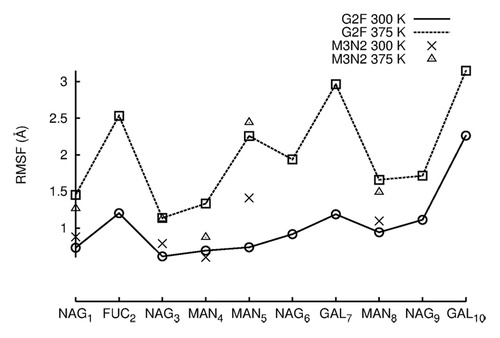
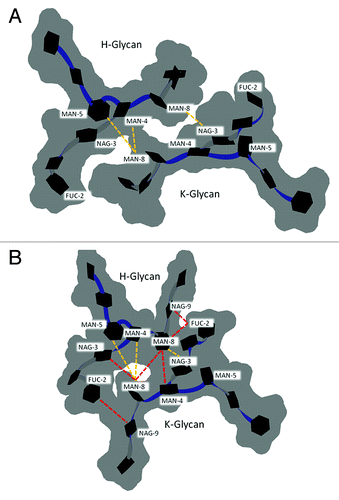
Figure 6. Glycan-glycan conformation for G2F in the (A) crystal structure of PDB entry 1HZH with GAL10 of chain K and FUC2 of chain H modeled in using MOE. (B) The preferred glycan-glycan conformation in 300 K simulations (see Materials and Methods). Carbohydrates (black hexagons) are represented using Twister and Paper Chain representations in VMD.Citation57,Citation58 Gray outlines show the surface of carbohydrates. Yellow dotted lines indicate inter-glycan contacts (identified at ≤ 4.5 Å for non-hydrogen atoms) between chains H and K in the crystal structure. Red dotted lines indicate inter-glycan contacts in the preferred glycan-glycan conformation that were formed during simulations at 300 K and are not formed in the crystal structure for 1HZH.