Figures & data
Figure 1 Kinetic binding analysis of XOMA 052 and XMA005. (A) Injection of human IL-1β over XOMA 052 captured by protein A/G. Data were collected in triplicate for six IL-1β concentrations ranging from 23 pM to 57.8 nM. (B) Three injections of 57.8 nM human IL-1β were followed for 4 h of dissociation and fit using Scrubber2 software to estimate a dissociation rate (kd) of ≤6.3 × 10−6 sec−1. (C) The trend in the residuals from the fit of the data in the early part of dissociation profile is not seen in experiments using aldehyde-coupled antibody and is thus believed to result from instability due to capture. The value for the dissociation rate was determined from the later portion of the long dissociations. The association rate of 1.7 × 106 M−1s−1 was determined using a global fit of the association curves with a simple 1:1 Langmuir binding model with the dissociation rate fixed at the value determined from the long dissociations. The fit of the data is shown as a solid red line, yielding an affinity of ≤4 pM. (D) Injection of mouse IL-1β over XOMA 052 captured by protein A/G. Data were collected in triplicate for six IL-1β concentrations ranging from 23 pM to 300 nM, except for the highest concentration, which was a singlicate injection. Data were fit globally with a simple 1:1 Langmuir binding model, yielding an affinity of 7 nM. Analysis of XOMA 052 (E) and XMA005 (F) binding to human IL-1β by Kinetic Exclusion Assay (KinExA). This solution-based method yields equilibrium binding constants (KD) of 300 and 240 fM with 95% confidence intervals of 115 to 742 and 70 to 722 fM, for XOMA 052 and XMA005, respectively.
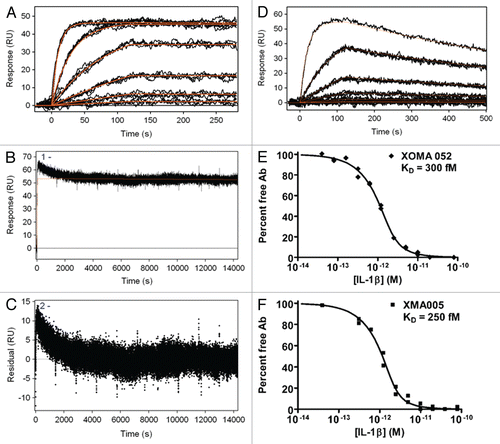
Figure 2 XOMA 052 cross-reacts with four species orthologs of IL-1β. Kinetic titration analysis of XOMA 052 binding to IL-1β from human (A), rhesus (B), rat (C) and mouse (D). Five concentrations of recombinant human, rhesus, rat and mouse IL-1β were injected in serial over XOMA 052 immobilized on a Biacore 2000 biosensor surface by aldehyde coupling. KD was determined by fitting data to a 1:1 Langmuir model using kinetic titration analysis with BIAevaluation software.
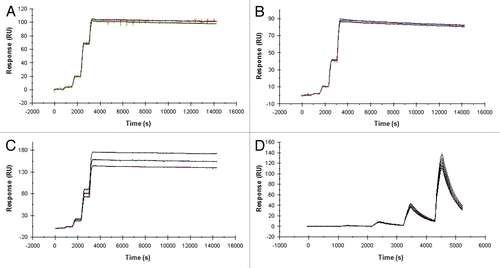
Figure 3 XOMA 052 epitope mapping. (A) The IL-1β PepSpot™ Peptide Array membrane probed with XOMA 052 reveals that XOMA 052 binds to peptide spots corresponding to amino acids 83–105 of the mature protein. (B) Alanine substituted peptides hybridized with XOMA 052. Sequences of the 16 peptides with the alanine substitution (in blue) are shown in the box below. Peptides 9–12 and 16 showed little or no binding by XOMA 052 (WT, wild type). (C) Sequence alignment of mature forms of mouse (m), human (h), rhesus (rh), rat (r) and rabbit (ra) IL-1β are shown. Residues that are identical in human, rhesus, rat and rabbit and differ in mouse are shown in bold and underlined. (D) Supernatants from wild type and six mutants of IL-1β (E64A, K65A, M95A, E96A, K97A and Q116E) were injected over XOMA 052 immobilized on a ProteOn XPR sensor chip. The fits of the off-rate data are shown as red lines. Mutants E96A, K97A and Q116E showed off-rates increased by 1,000-fold. (E) Sensorgrams of wild type and IL-1β mutants binding to sRI show that the mutant proteins were expressed and folded properly.
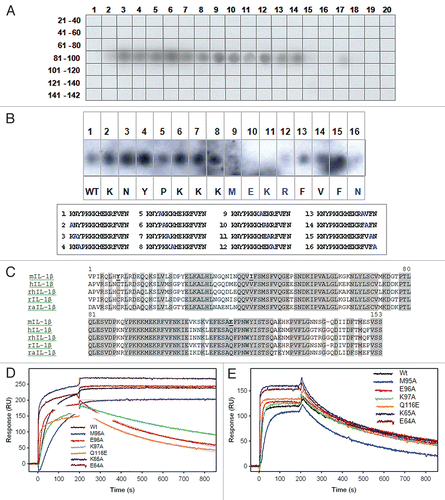
Figure 4 XOMA 052 epitope as predicted by PepSpot™ and alanine scan analyses. Figure shows the ribbon representation of the structure for the human IL-1β/IL-1RI complexCitation22 using Pymol visualization software (DeLano Scientific LLC, San Carlos, CA). Receptor domains I, II and III are depicted in green, yellow and orange respectively and IL-1β depicted in blue. The side chains of E96, K97 and Q116 identified as critical for binding to XOMA 052 are shown in red. Center: front view, left: a 90° rotation view to the left, right: a 90° rotation view to the right.
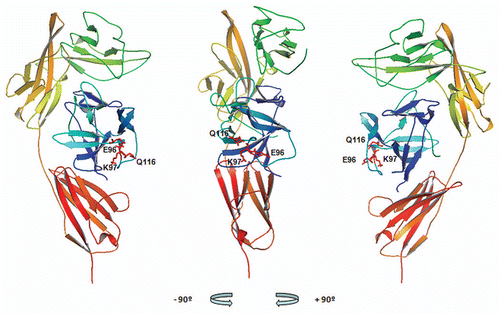
Figure 5 XOMA 052 neutralizes human IL-1β activity in vitro. (A) Inhibition of IL-1β-induced IL-6 release in MRC5 human fibroblast cells. Cells were stimulated with 5.8 pM of IL-1β in the presence of XMA005, XOMA 052, anakinra or a control IgG2 Ab. XMA005 and XOMA 052 had an IC50 of 4.9 and 4.4 pM respectively, while anakinra had an IC50 of 45 pM. (B) Inhibition of IL-1β induced IL-8 expression in a human whole blood assay. Whole blood was stimulated with 100 pM of IL-1β in the presence of XOMA 052, anakinra or a control IgG2 Ab. XOMA 052 has an IC50 of 28 pM, while recombinant human IL-1Ra anakinra has an IC50 of 603 pM.
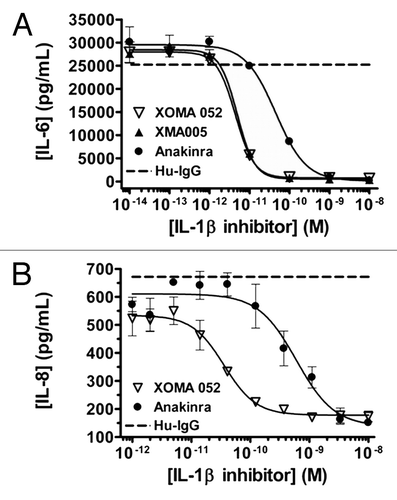
Figure 6 XOMA 052 inhibits IL-6 expression in IL-1β treated mice. Mice (n = 8 per treatment group) were injected with XMA005 parent, XOMA 052 or IgG control and treated 24 h later with 1 µg/kg of recombinant human IL-1β as described in Materials and Methods. Two hours later mice were sacrificed and neutralization activity of the antibodies was calculated by measuring the levels of IL-6 in the mouse serum. Percent neutralization was calculated using the mean of the IgG control treatment group as 100%. Both antibodies inhibited IL-6 stimulation in a dose-dependent manner with comparable potency. Values are mean percent neutralization, and error bars show standard error of the mean. The difference between XMA005 and XOMA 052 is not significant as measured by unpaired two-tailed t-test (p = 0.17, 0.41 and 0.36 for 0.15, 0.05 and 0.015 mg/kg doses, respectively).
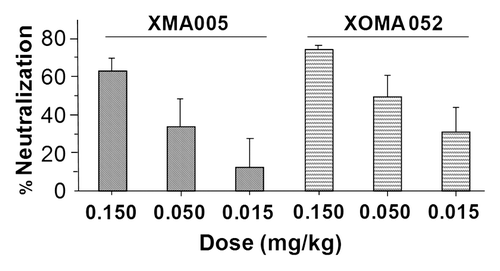
Figure 7 XOMA 052 blocks inflammation in a model of acute gout. (A) Number of infiltrating neutrophils in the peritoneal lavage, 6 h after MSU injection. (B) Number of infiltrating macrophages in the peritoneal lavage, 6 h after MSU injection. n = 6 mice/group. *p < 0.05 for XOMA 052 10 mg/kg vs. IgG2 control or anakinra vs. PBS using an unpaired t-test.
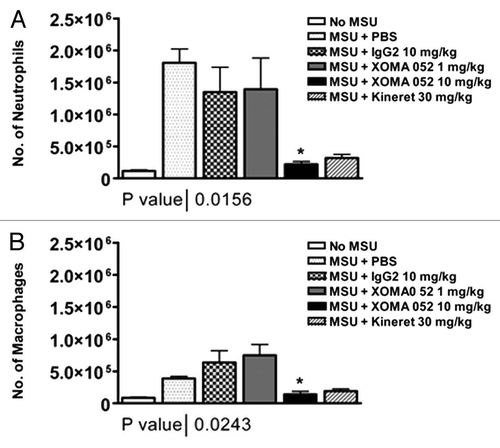
Table 1 Comparative binding affinities and kinetics of XOMA 052 binding to IL-1β from different species determined by Biacore
Table 2 Effect of single amino acid substitutions of human IL-1β on binding to sRI and XOMA 052