Figures & data
Figure 1 Schematic description of artificial fouling of MabSelect SuRe resin in PreDictor plates. Steps 1, 3 and 5 show equilibration or wash with PBS (20 mM phosphate, 0.15 M NaCl, pH 7.4). In step 2, which was repeated four times, E. coli lysate spiked with polyclonal IgG was added to the resin and incubation was done for 45 min. In step 4, a fouling agent composed of 2.9 M ammonium sulfate, 0.6 M phosphoric acid, pH 2.5 was added, followed by incubation overnight in order to accelerate the fouling of the resin. After a subsequent wash, the bound IgG was eluted with 0.1 M sodium citrate pH 3.0. The resin in the wells was mixed at every step and liquid was removed by centrifugation between each step. H2O, ultrapure water.
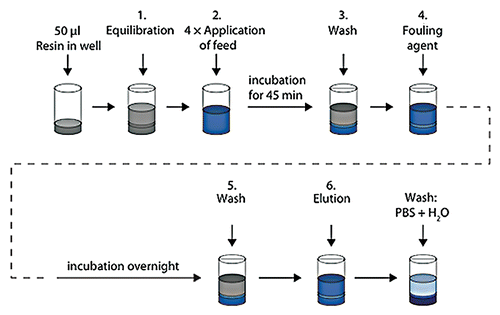
Figure 2 Evaluation of cleaning conditions. Different cleaning conditions were evaluated by incubation of the fouled MabSelect SuRe resin in cleaning chemicals and sequences of cleaning steps. An incubation time of 15 min was used, which corresponds to the CIP contact time in a column. Between each cleaning step in a sequence, the resin was washed with 300 µL PBS followed by wash with 300 µL H2O. In each step, mixing was done and centrifugation was used for liquid removal between each step.
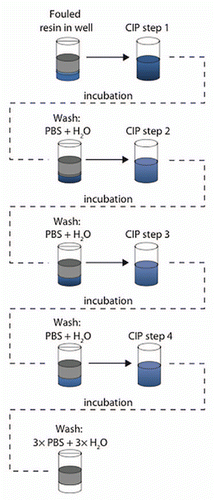
Figure 3 Reduced SDS-PAGE (Deep Purple™ stained) for analysis of proteins remaining on the artificially fouled MabSelect SuRe resin after cleaning with different agents. The control sample, which was washed with PBS (lane 1), showed that there was a large amount of proteins on the artificially fouled resin. NaOH proved to be very effective in cleaning, and increasing NaOH-concentration resulted in increasing cleaning efficiency (lanes 2–4). 0.1 M NaOH + 1.0 M NaCl (lane 7) resulted in a significant decrease in cleaning efficiency compared to 0.1 M NaOH without salt (lane 2).
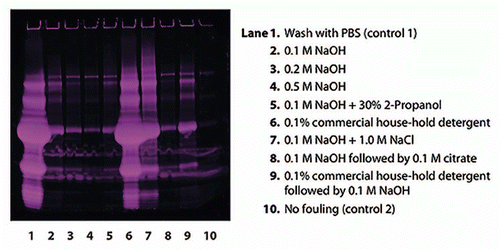
Figure 4 Comparison of samples prepared in a filter plate to samples prepared in a collection plate. Artificially fouled PreDictor MabSelect SuRe was cleaned using: (1) PBS (20 mM phosphate, 0.15 M NaCl, pH 7.4) (control 1), (2) Proprietary CIP solution, (3) 0.15 M phosphoric acid, pH 1.6, (4) 6.0 M Guanidine hydrochloride, (5) 0.1 M NaOH + 1.0 M NaCl, (6) 0.1 M NaOH, (7) 0.5 M NaOH, (8) PBS, no fouling (control 2). The relative protein concentration on the resin was measured using chip electrophoreses (n = 4, mean ± SD).
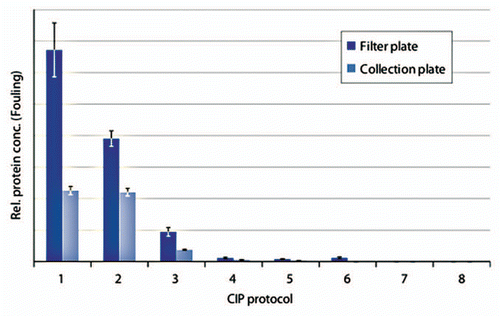
Figure 5 Evaluation of occurrence of cross-contamination between wells during resin transfer using centrifugation. MabSelect SuRe resin in every second column in the first half of a PreDictor plate and in every second row in the second half of the PreDictor plate was colored using bromophenol blue (top). After liquid evacuation the drained resin was transferred to a collection plate using centrifugation (bottom).
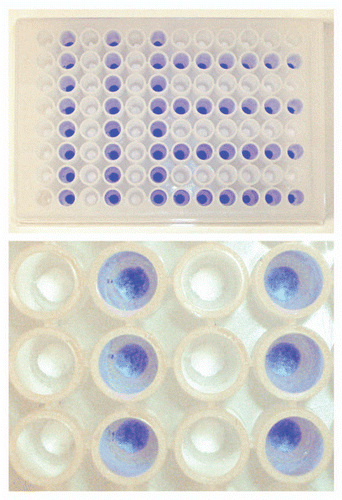
Figure 6 Schematic description of fouling of MabSelect SuRe resin in PreDictor plates by repeated bind-elute cycles. Steps 1 and 3 show equilibration or wash with PBS (20 mM phosphate, 0.15 M NaCl, pH 7.4). In step 2, harvested cell culture fluid with mAb was added to the resin and incubation was done for 30 min. In step 4, the bound mAb was eluted with 0.1 M sodium citrate pH 3.5. Steps 1–4 were repeated ten times, corresponding to ten chromatography cycles. In each step, mixing was done and centrifugation was used for liquid removal between each step.
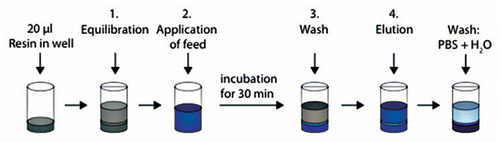
Figure 7 Chip electrophoresis analysis of residual impurities on MabSelect SuRe after cleaning. The Caliper software generates virtual slab gels (left) and electropherograms (right). Low intensity lane in the virtual slab gel, and corresponding electropherogram, representing the most efficient CIP protocol in this figure, are highlighted. The main protein impurity on the fouled MabSelect SuRe resin was mAb; the large size molecular peak in B5 is most likely un-reduced antibody.
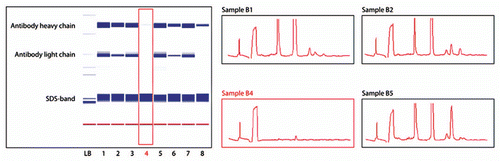
Figure 8 Remaining impurities on fouled MabSelect SuRe resin after cleaning with 32 different protocols including one control (n = 3, mean ± SD). Detailed information about the composition of the CIP protocols is found in . Low protein concentration on the resin corresponded to efficient cleaning and vice versa. CIP protocol 11 is cleaning with 0.1 M NaOH. In CIP protocols 12–20, 0.1 M NaOH was combined with acidic steps, solvent, alcohol or chaotropic agents. CIP 21–23 corresponded to 0.3–0.5 M NaOH with/without acidic cleaning. In CIP 24–29 the resin was cleaned with reducing agent followed by 0.1 M NaOH with or without acidic cleaning and CIP 30 and 31, reducing agent was followed by 0.5 M NaOH. In CIP 32, the reversed order, i.e., 0.1 M NaOH followed by 100 mM 1-thioglycerol was used.
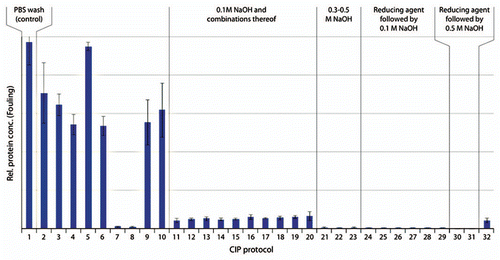
Table 1 Thirty-two different cleaning protocols including one control (wash with PBS) were evaluated for cleaning efficiency of fouled MabSelect SuRe resin