Figures & data
Figure 1 Variable domain sequences of mouse and humanized variants of anti-FGFR4. The amino acid sequences of mouse LD1 and humanized variants hLD1.vB and hLD1.v22 are aligned with the (A) human kappa I (huKI) and (B) human VH subgroup III (huIII) variable domain frameworks used in trastuzumab. Differences are highlighted in yellow and positions are numbered according to Kabat. Hypervariable regions that were grafted from mouse LD1 into the human variable Kappa I and subgroup III consensus frameworks were selected based on a combination of sequence, structural and contact CDR definitionsCitation16 and are boxed. Three vernier positions in the light chain were altered to restore affinity during humanization; these positions are not expected to be surface exposed.
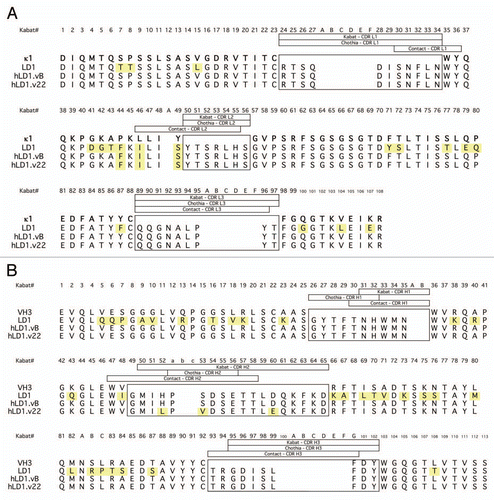
Figure 2 Pharmacokinetics and Distribution of anti-FGFR4 variants. (A) Comparison of the binding of chLD1 and hLD1.vB to FGFR4 using the FGFR4 ELISA. (B) Comparison of day 16 tumor volumes of chLD1, hLD1.vB and vehicle in an HUH7 human hepatocelluar carcinoma xenograft model in CRL nu/nu mice. Antibodies were administered at doses of 30 mg/kg twice weekly (10 mice per group). Only chLD1 was effective at reducing tumor growth relative to the PBS control (p value = 0.014) while hLD1.vB was not significantly effective (p value = 0.486). (C) Pharmacokinetics of chLD1 (open symbols) and hLD1.vB (closed symbols). NCR nude mice were IV administered 1 (dashed lines) or 20 (solid lines) mg/kg doses and samples were analyzed using the FGFR4 ELISA. Similar results were obtained using the IgG ELISA (not shown). (D) Tissue distribution of 125I-chLD1 and 125I-hLD1.vB in NCR nude mouse. Mice were administered either 125I-chLD1 (black bars) or 125I-hLD1.vB (open bars) and the percent of injected dose per gram of tissue (% ID/g) was determined at 2 h post-dose as described in Methods.
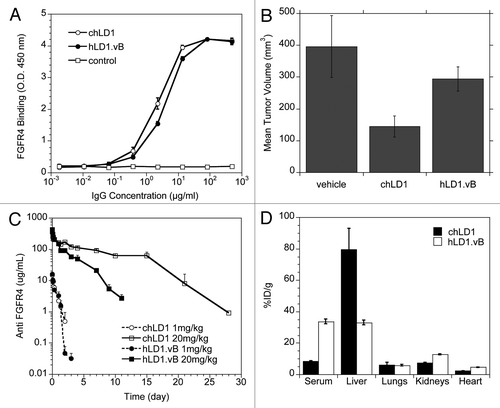
Figure 3 Identification of an interaction between hLD1.vB and mouse C3d. (A) Detection of chLD1 (black bar) and hLD1.vB (white bar) following incubation for 48 h in PBS/BSA or NCR nude mouse, rat, human and cynomolgus monkey plasma. Percent recovery was determined using the FGFR4 ELISA. (B) Plasma binding analysis of 125I-chLD1 (solid line) and 125I-hLD1.vB (dashed line). The traces are off-set; a dot indicates the position of the 150 kDa peaks. The antibodies were incubated in mouse plasma for 0 and 48 h followed by analysis using size exclusion HPLC. For incubations in PBS/BSA, human plasma or cynomolgus monkey plasma see Supplemental Figure 1. All incubations gave rise to a peak at the expected 150 kDa corresponding to IgG. Higher molecular weight peaks were only observed in mouse plasma samples containing hLD1.vB. The initial time point revealed additional peaks at ca. 270 and ca. 550 kDa while at 48 h, only the 270 kDa peak was observed. The high molecular weight peaks were not observed when hLD1.vB/mouse plasma incubations were performed at pH 4, indicating that the presence of these high molecular weight peaks was pH dependent (Sup. Fig. 1). (C) Immunoprecipitation of mouse plasma. chLD1 and hLD1.vB were incubated in mouse plasma for 24 h at 37°C and analyzed by size-exclusion HPLC. Protein-G beads were then added to fractions followed by SDS-PAGE analysis. A band at ca. 37 kDa was detected in fractions corresponding to the 270 kDa peak present in the hLD1.vB/mouse plasma sample (lane 3), but not in samples from cynomolgus monkey or human plasma or PBS/BSA (Sup. Fig. 1). This band was not observed in mouse plasma alone (lane 4) or mouse plasma incubated with chLD1 (lane 2). The protein molecular weight marker was run in lane 1. (D) MS/MS sequence coverage of mouse C3 obtained from immunoprecipitation with hLD1.vB. The sequence for mouse C3 is shown and identified peptides are highlighted in red; the region encoding C3d is underlined.
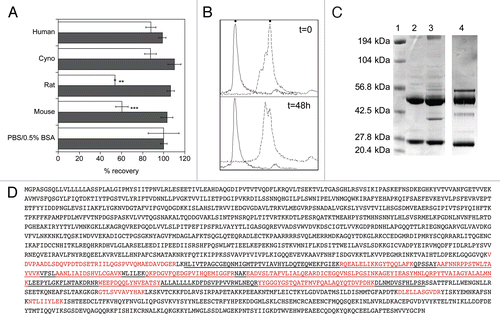
Figure 4 An affinity matured anti-FGFR4 variant lacks C3d binding. (A) Detection of FGFR4 binding of chLD1, hLD1.vB and hLD1.v22 incubated 16 h in NCR nude (black bar), C3 wt (open bar) or C3 ko (shaded bar) mouse serum prior to assessment using the FGFR4 ELISA. Samples were normalized to identical samples incubated in PBS/0.5% BSA. (B) Lack of mouse C3d immunoprecipitation by hLD1.v22. Immunoprecipitation from NCR nude mouse plasma using chLD1 (lane 2), hLD1.vB (lane 3) and hLD1.v22 (lane 4) was analyzed by SDS-PAGE. The band at ∼37 kDa was only detected in the hLD1.vB/mouse plasma sample. The protein molecular weight marker was run in lane 1.
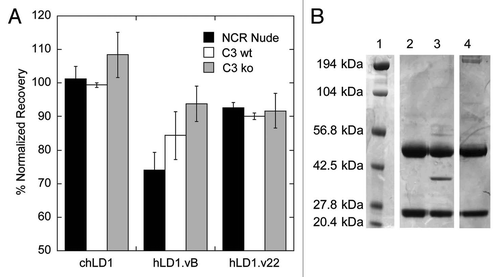
Figure 5 A loss in C3d binding restores Pharmacokinetics and Efficacy. (A) Pharmacokinetic analysis of chLD1 (open symbols) and hLD1.vB (solid symbols) in C3 wt (solid lines) and C3 ko (dashed lines) mice. Antibodies were IV administered at 20 mg/kg; their concentration in blood was monitored using the FGFR4 ELISA. The clearance of hLD1.vB in C3 ko mice is similar to that of chLD1 in both C3 ko and C3 wt mice. (B) Pharmacokinetic analysis of chLD1 (open circles), hLD1.vB (solid circles) and hLD1.v22 (solid diamond) in NCR nude mice. Antibodies were IV administered at 20 mg/kg; their concentration in blood was monitored using the FGFR4 ELISA. The clearance of hLD1.v22 is similar to that of chLD1. (C) Comparison of chLD1 (open circle), hLD1.vB (solid circle) and hLD1.v22 (solid diamond) in an HUH7 human HCC xenograft model in CRL nu/nu mice. Antibodies were administered at 30 mg/kg weekly (10 mice per group) and tumor volumes were monitored for 4 weeks. At day 21 chLD1 and hLD1.v22 (p value = 7 × 10−7 and 3 × 10−5, respectively) were effective at reducing tumor growth relative to the PBS control (open square) while hLD1.vB was only marginally effective (p value = 0.011).
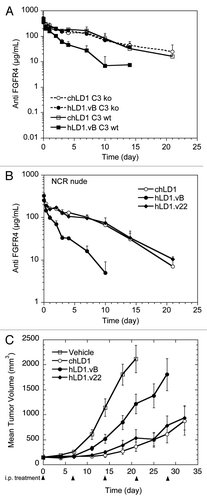
Table 1 Binding kinetics of anti-FGF.R4 variants