Figures & data
Table 1. Examples of functional epitope mapping through phage display-based antigen surface scanning
Figure 1. Schematic representation of the phage display-based antigen surface scanning platform for epitope mapping. The first step is successful phage display of the target antigen. A second challenge is the identification of a putative recognized antigenic region, which can be defined through several approaches. Once the candidate region is identified, it should be deeply explored, through extensive site-directed or combinatorial mutagenesis, in order to decipher the functional map of the epitope. Subsequent grafting experiments can confirm the identity of the functional epitope. All the procedures that could take advantage of the high throughput potential of phage display are enclosed in boxes.
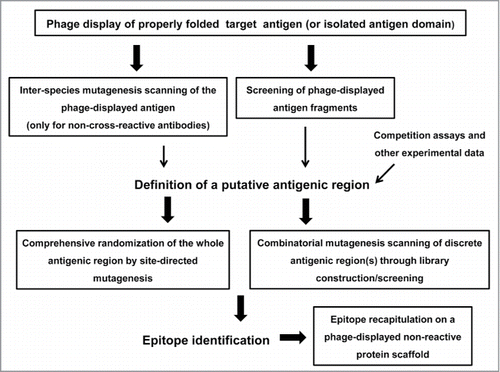
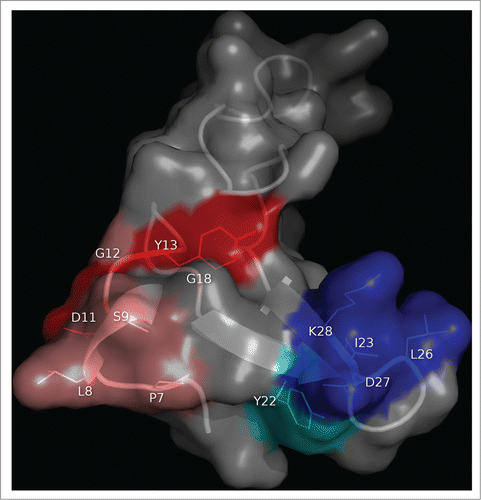
Figure 2. Functional dissection of the structural epitope recognized by cetuximab on EGF receptor. EGFR domain III (PDB code 1YY9) is shown as a cartoon with semi-transparent surface. Side chains of residues that contribute to binding (according to the site-directed mutagenesis scanning on the phage-displayed molecule) are represented with sticks and colored as follows. Critical residues that cannot be replaced (or can only be substituted by residues sharing their physicochemical properties) are highlighted in red. Other residues that can be replaced by some, but not all, amino acids (with no obvious shared features among them) are shown in orange. Magenta indicates that the corresponding residue can only be substituted by amino acids with smaller side chains, implying that its functional contribution is limited to the lack of steric hindrance. The rest of cetuximab structural epitope is represented in cyan. The figure was generated with Pymol.
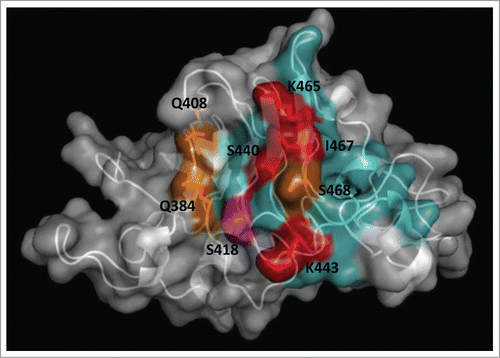
Figure 3. Clusters of residues identified by combinatorial mutagenesis as the epitopes recognized by 2 monoclonal antibodies against EGF. The antigen (PDB code 1IVO_C) is represented as a cartoon with semi-transparent surface. Residues recurrently found among EGF mutated variants selected from libraries on CB-EGF.1 or CB-EGF.2 mAbs were considered to contribute to the formation of each epitope. The corresponding side chains are represented with lines and colored as described below. A closer examination of their relative abundances allowed the subsequent classification of relevant residues as functional contributors/major functional contributors. Residues belonging to each of these categories are highlighted in salmon/red (for CB-EGF.1 mAb) and cyan/blue (in the case of CB-EGF.2). The figure was generated with Pymol.