Figures & data
Figure 1. Strategy for monoclonal antibody development by naïve antibody phage display library panning and B cell hybridoma technique.
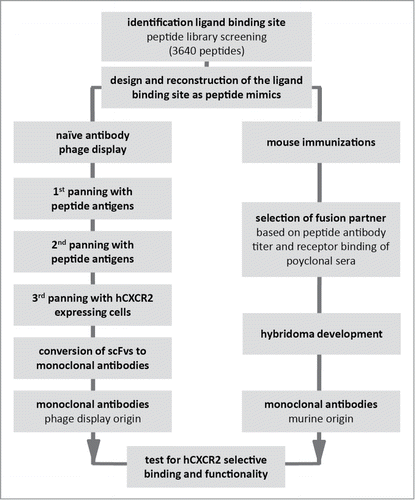
Figure 2. Identification of IL-8 binding sites on CXCR2. (A) Binding of IL-8 to array of > 3,600 different peptide constructs. The peptide arrays were challenged with biotinylated IL-8 and detected with streptavidin-HRP. (B) Cartoon of CXCR2. Seven transmembrane domains (cylinders) are connected by 3 intra cellular loops (not labeled) and 3 extracellular loops (ECL1, ECL2 and ECL3) with 2 disulfide bonds (solid lines), between ECL3 and N-terminus, and between ECL1 and ECL2, respectively. Black dots represent the inferred binding positions of IL-8. (C) Peptide reconstruction of the extracellular regions of hCXCR2. N-terminus (DSFEDFWKGEDLSNYSYSSTLPPFLLDAAPCEPESLEINK) (black structure) and ECL3 (DTLMRTQVIQETCERRNHIDR) (dark gray arch) are connected by a disulfide bond (antigen A) or a CLIPS™ moiety (antigen B). The biotin moiety (gray square), for antibody phage display library panning, is located at the C-terminal side of the N-terminus.
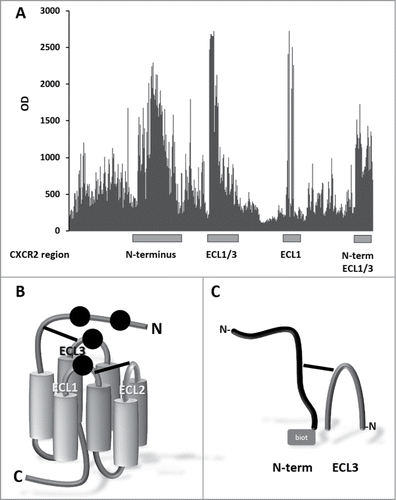
Table 1A. scFvs isolated from panning naïve antibody phage display libraries
Table 1B. Antibodies from mouse hybridomas, elicited with antigen A
Figure 3. Top Panel: Inhibition of IL-8 (A and C) and Gro-α (B and D) induced ß-arrestin recruitment in hCXCR2-HEK cells (Tango™ GPCR Assay System, Invitrogen) by IgGs converted from scFvs (A and B): 4138.51 (open square), 4081.73 (solid circle), 4138.73 (filled diamond), 4138.06 (asterisk), 4138.27 (open triangle), 4138.39 (open circle), 4138.14 (solid triangle) 4138.54 (solid star), and 4138.40 (open diamond). Murine IgGs (C and D): mAb27 (solid diamond), mAb5 (solid circle), mAb9 (solid square), mAb17 (open square), mAb7 (open circle), mAb21 (open diamond) and mAb19 (solid triangle). Each curve is based on 2 independent measurements except for panel A which is based on 4 independent measurements. The error bars represent the standard deviations. Bottom panel: Using GraphPad Prism software (Software MacKiev) the top values of each dataset were constrained at 100% and the bottom values of each dataset were shared. In all panels the Hill slopes were constrained at -1.000. IgGs converted from scFvs (A and B): 4138.51 (a), 4081.73 (b), 4138.73 (c), 4138.06 (d), 4138.27 (e), 4138.39 (f), 4138.14 (g) 4138.54 (h), and 4138.40 (i). Murine IgGs (C and D): mAb27, mAb5, mAb9, mAb17, mAb7, mAb21 and mAb19. Each curve is based on 2 independent measurements except for panel A which is based on 4 independent measurements.
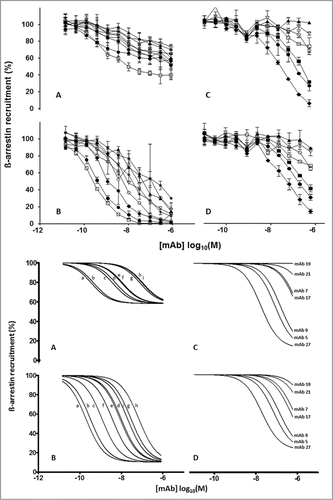
Table 2. Binding to native CXCR2 and functional testing of recombinant IgGs (converted from scFvs) and murine monoclonal antibodies
Table 3. Epitope mapping of recombinant IgGs (converted from scFv) and murine monoclonal antibodies
