Figures & data
Figure 1 hFcRn binding and blocking activity of anti-hFcRn antisera. (A) Representation of the construct design lacking the cytoplasmic targeting domain (left). Confocal images of HeLa cells transiently transfected with the hFcRn-GFP after incubation with hIgG at the indicated pH, fixed and stained with goat anti-hIgGAF568 (right). (B) Flow cytometric data of 293hFcRn-GFP cells incubated with hIgGAF647 at pH 6.0 (left) or 7.2 (right). (C) Representative results from evaluation of antisera from mice primed with hFcRn. Binding of the antisera to hFcRn-GFP was detected with goat anti-mouse IgG-PE at pH 7.2 (left). Functional blockade of hIgG binding to hFcRn was determined by the ability to inhibit the binding of hIgGAF647 at pH 6.0 (right). Values within scatter plots represent the AF647 mean fluorescence intensity (MFI) of each hFcRn-GFP+-gated cell population. Data are representative of at least two experiments.
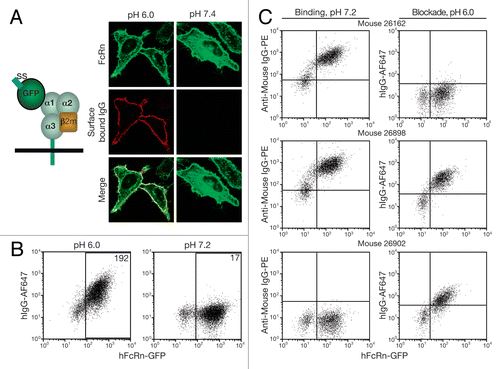
Figure 2 Anti-hFcRn mAb specificities and relative affinities. 293hFcRn-GFP, 293mFcRn-GFP, or control 293 cells were incubated with AF647-labeled anti-hFcRn mAbs (A) at a fixed concentration of 200 ng/106 cells to determine specificity or (B) with a range of concentrations to assess relative mAb affinities for hFcRn. Values within scatter plots represent the AF647 MFIs of each hFcRn-EGFP+-gated cell population. Data are representative of at least two experiments.
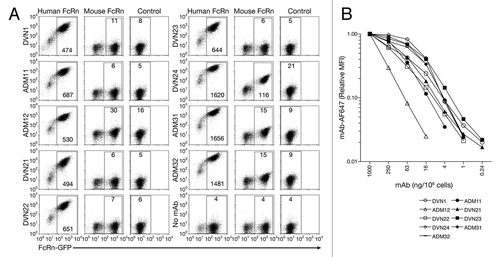
Figure 3 Immunofluorescent detection of hFcRn (red channel) in kidney tissue sections from (A) hFcRn transgenic mice, (B) hFcRn transgenic mice costained for proximal convoluted tubule (blue channel) and podocytes (green channel), (C) wild-type B6 mice costained for proximal convoluted tubule and podocytes, (D) human tissue, (E) human tissue costained for proximal convoluted tubule and podocytes, and (F) human tissue showing only proximal convoluted tubule and podocytes (GM, Glomerulus, PCT, proximal convoluted tubules).
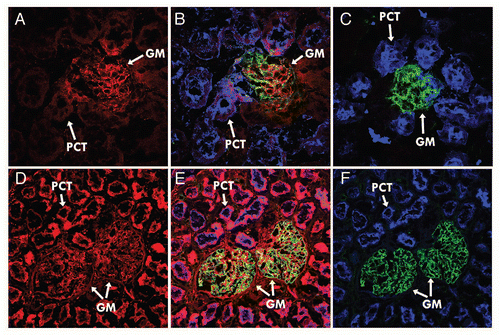
Figure 4 hFcRn blocking activity. 293hFcRn-GFP cells were incubated with a range of concentrations of DVN1 (○), ADM11 (●), DVN21 (▴), DVN22 (□), DVN23 (■), DVN24 (◊), ADM31 (◆) or ADM32 (−) at pH 6, then stained for functional binding with (A) 20 µg/mL hIgGAF647, or (B) 200 µg/mL HSAbiotin followed by streptavidin-PE. Nonfunctional binding was indicated by the AF647 or PE MFIs of cells stained with labeled ligands at pH 7.2 (X). (C) Scatter plots of 293hFcRn-GFP cells that were untreated at pH 6.0 (upper left) or at pH 7.2 (upper right), exposed to 200 ng/mL of DVN24 (lower left) or ADM11 (lower right) at pH 6.0, and then stained for functional binding of hIgGAF647 at the corresponding pH. (D) Scatter plots of 293hFcRn-GFP cells that were untreated at pH 6.0 (upper left) or at pH 7.5 (upper right), exposed to 200 ng/mL of ADM31 (lower left) or ADM11 (lower right), and then stained for functional binding with HSA-biotin and counterstained with streptavidin-PE at the corresponding pH. AF647 or PE MFI values of the hFcRn-GFP+-gated cell populations are indicated. Data are representative of three independent experiments.
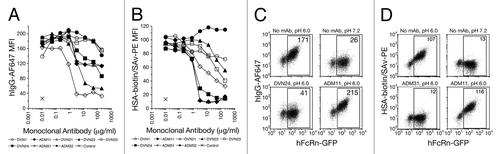
Figure 5 Blockade of IgG by anti-hFcRn mAbs in vivo. (A) B6 Tg276 mice were co-injected ip with 100 µg each of hIgG, mIgG1 (1B7.11) and mIgA (2F11–10) as tracers. The mice were then injected with 1,000 µg (◊), 300 µg (◆), 100 µg (△), 30 µg (▴) or 0 µg (□) of DVN24 or with 1,000 µg (X) of a control IgG2a mAb into groups of 3 mice on days 2, 3 and 4 d after tracer injection. (B) B6 Tg276 mice (n = 3) were co-injected ip with 100 µg hIgG and 1 mg of HSA tracers followed by injections with 1 mg doses of ADM32 (−), DVN24 (◊) or isotype matched mIgG2a (X). (C) B6 Tg276 mice (n = 4–5) were injected with 100 µg hIgG tracer and then treated on days 5–7 or 10–12 with 1 mg of DVN24 (◊) or mIgG2a (X). Significant differences (A–C) are indicated as *p < 0.05 and **p < 0.01 comparing 1,000 µg injections of DVN24 with the control isotype matched mIgG2a. (D) B6 WT mice (n = 8) were injected ip with 200 µg of mIgG1 (1B7.11) as a tracer, and then injected ip on days 4–6 with 1 mg doses of DVN24 (◊), ADM31 (◆) or vehicle only (X). Tracer plasma concentrations were determined by ELISA and plotted either as percent remaining as compared with the first time point plasma concentrations or as plasma concentrations ±SD. Significant differences (D) are indicated as **p < 0.01 comparing 1,000 µg injections of DVN24 with the vehicle only control.
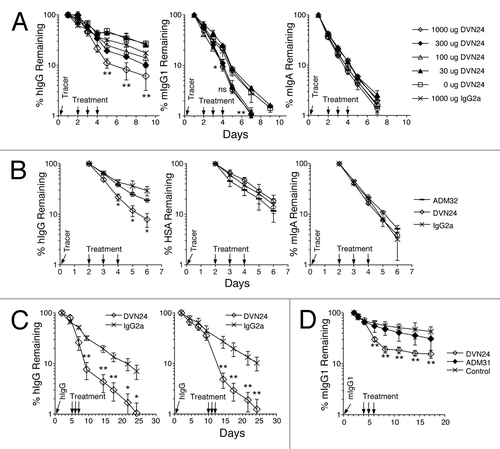
Table 1 Competitive binding of anti-hFcRn mAbsTable Footnotea
Table 2 Summary of anti-hFcRn mAb characteristics