Figures & data
Figure 1 pAMα1 and amplification of tetracycline resistance. (A) pAMα1 is shown as being originally generated by the formation of a cointegrate structure through a specific and reversible recombination mechanism involving relaxase operating on the oriT sites of two smaller plasmids pAMα1Δ1 and pAMα1Δ2. (B) Amplification generating multiple copies of the tet determinant via recombination between the two “recombination sequences” RS1 and RS2. The first event involves relaxase and could, for example, bring about excision of the segment containing tet and its reinsertion into an intact pAMα1 copy. Other mechanisms, not mutually exclusive, are possible (see text. Subsequent steps can involve host-directed homologous recombination.
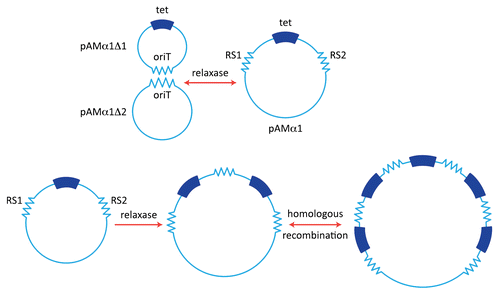
Figure 2 Tn916 behavior and map. (A) The transposon is shown as excising and generating a circular, plasmid-like but nonreplicative intermediate, which can then insert into another site in the genome (on chromosome or a plasmid) or transfer by a plasmid-like process to a recipient cell. (B) A simplified view of the transposon showing the general locations of determinants involved in excision/insertion, tetracycline resistance and conjugation.
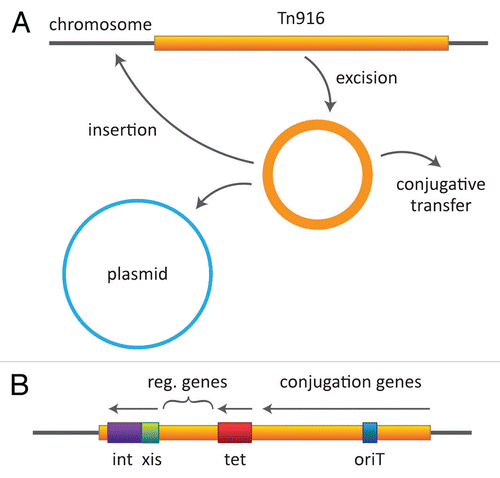
Figure 3 Map of pAD1. The related functions encoded on the plasmid are noted in different colors with the various determinants indicated. A detailed description of the specific regions can be found in reference Citation131 (redrawn from ref. Citation131).
Figure 4 Map of the replication and partitioning region of pAD1. Two clusters of iterons (TAGTARRR) are shown between repA and repBC. Isolated iteron sequences are also located within the promoters of repA and repBC. RepC binds cooperatively to iteron sequences; in the presence of ATP, RepB also binds. RepC binding to the isolated iterons probably affects expression of repBC and repA. A “folded” conformation may be stabilized by the binding and effect interaction between the opposing promoters. Phase variation resulting from changes in the number of iterons could affect this interaction and modulate expression of RepBC and RepA (redrawn from ref. Citation131).
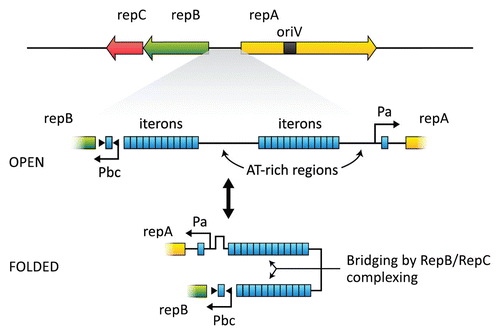
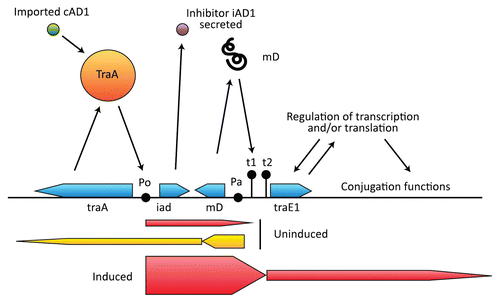
Figure 5 pAD1 region that regulates conjugation. The negative regulator TraA binds to the promoter P0 and dissociates when bound to the pheromone cAD1. A limited amount of transcription occurs from P0 to the terminator t1 in the uninduced state, allowing expression of the inhibitor iAD1. Pa is the promoter for mD, an RNA that enhances transcription termination at t1. Pa is also the promoter for traA. Induction results in transcription through t1 and t2 and into traE1, which encodes a positive regulator for conjugation-related determinants as well as itself.